Protein aggregation is a consequence of the dormancy-inducing membrane toxin TisB in Escherichia coli
- PMID: 39377584
- PMCID: PMC11575346
- DOI: 10.1128/msystems.01060-24
Protein aggregation is a consequence of the dormancy-inducing membrane toxin TisB in Escherichia coli
Abstract
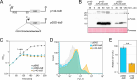
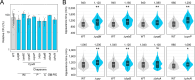
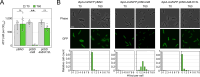
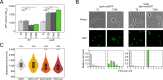
Bacterial dormancy is a valuable strategy to survive stressful conditions. Toxins from chromosomal toxin-antitoxin systems have the potential to halt cell growth, induce dormancy, and eventually promote a stress-tolerant persister state. Due to their potential toxicity when overexpressed, sophisticated expression systems are needed when studying toxin genes. Here, we present a moderate expression system for toxin genes based on an artificial 5' untranslated region. We applied the system to induce expression of the toxin gene tisB from the chromosomal type I toxin-antitoxin system tisB/istR-1 in Escherichia coli. TisB is a small hydrophobic protein that targets the inner membrane, resulting in depolarization and ATP depletion. We analyzed TisB-producing cells by RNA-sequencing and revealed several genes with a role in recovery from TisB-induced dormancy, including the chaperone genes ibpAB and spy. The importance of chaperone genes suggested that TisB-producing cells are prone to protein aggregation, which was validated by an in vivo fluorescent reporter system. We moved on to show that TisB is an essential factor for protein aggregation upon DNA damage mediated by the fluoroquinolone antibiotic ciprofloxacin in E. coli wild-type cells. The occurrence of protein aggregates correlates with an extended dormancy duration, which underscores their importance for the life cycle of TisB-dependent persister cells.
Importance: Protein aggregates occur in all living cells due to misfolding of proteins. In bacteria, protein aggregation is associated with cellular inactivity, which is related to dormancy and tolerance to stressful conditions, including exposure to antibiotics. In Escherichia coli, the membrane toxin TisB is an important factor for dormancy and antibiotic tolerance upon DNA damage mediated by the fluoroquinolone antibiotic ciprofloxacin. Here, we show that TisB provokes protein aggregation, which, in turn, promotes an extended state of cellular dormancy. Our study suggests that protein aggregation is a consequence of membrane toxins with the potential to affect the duration of dormancy and the outcome of antibiotic therapy.
Keywords: antibiotics; dormancy; protein aggregation; toxin-antitoxin systems; type I toxins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

