Optimizing a CRISPR-Cas13d Gene Circuit for Tunable Target RNA Downregulation with Minimal Collateral RNA Cutting
- PMID: 39377757
- PMCID: PMC11494644
- DOI: 10.1021/acssynbio.4c00271
Optimizing a CRISPR-Cas13d Gene Circuit for Tunable Target RNA Downregulation with Minimal Collateral RNA Cutting
Abstract
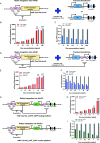
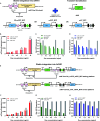
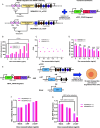
The invention of RNA-guided DNA cutting systems has revolutionized biotechnology. More recently, RNA-guided RNA cutting by Cas13d entered the scene as a highly promising alternative to RNA interference to engineer cellular transcriptomes for biotechnological and therapeutic purposes. Unfortunately, "collateral damage" by indiscriminate off-target cutting tampered enthusiasm for these systems. Yet, how collateral activity, or even RNA target reduction depends on Cas13d and guide RNA abundance has remained unclear due to the lack of expression-tuning studies to address this question. Here we use precise expression-tuning gene circuits to show that both nonspecific and specific, on-target RNA reduction depend on Cas13d and guide RNA levels, and that nonspecific RNA cutting from trans cleavage might contribute to on-target RNA reduction. Using RNA-level control techniques, we develop new Multi-Level Optimized Negative-Autoregulated Cas13d and crRNA Hybrid (MONARCH) gene circuits that achieve a high dynamic range with low basal on-target RNA reduction while minimizing collateral activity in human kidney cells and green monkey cells most frequently used in human virology. MONARCH should bring RNA-guided RNA cutting systems to the forefront, as easily applicable, programmable tools for transcriptome engineering in biotechnological and medical applications.
Keywords: CRISPR; Cas13d; RNA-targeting; collateral activity; gene downregulation; synthetic gene circuit.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

