The Evolutionary Forest of Pancreatic Cancer
- PMID: 39378050
- PMCID: PMC11803399
- DOI: 10.1158/2159-8290.CD-23-1541
The Evolutionary Forest of Pancreatic Cancer
Abstract
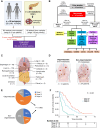
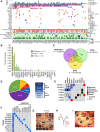
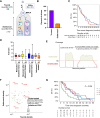
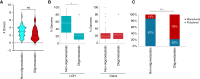
Although the pancreatic cancer genome has been described, it has not been explored with respect to stages of diagnosis or treatment bottlenecks. We now describe and quantify the genomic features of PDAC in the context of evolutionary metrics and in doing so have identified a novel prognostic biomarker.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
H. Zhang reports other support from Valar Labs outside the submitted work. A.P. Makohon-Moore reports grants from the NCI, the American Cancer Society, the Scott Mackenzie Foundation, the American Association for Cancer Research, and the New Jersey Health Foundation during the conduct of the study. A. Zucker reports grants from Ruth L Kirschstein T32 MD/PD Training Grant outside the submitted work. N.D. Socci reports grants from NIH during the conduct of the study. C.A. Iacobuzio-Donahue reports other support from Bristol Myers Squibb outside the submitted work. No disclosures were reported by the other authors.
Figures



References
-
- Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

