Somatic instability of the FGF14-SCA27B GAA•TTC repeat reveals a marked expansion bias in the cerebellum
- PMID: 39378335
- PMCID: PMC11969470
- DOI: 10.1093/brain/awae312
Somatic instability of the FGF14-SCA27B GAA•TTC repeat reveals a marked expansion bias in the cerebellum
Abstract
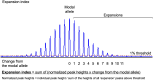
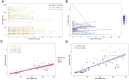
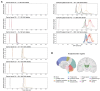
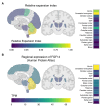
Spinocerebellar ataxia 27B (SCA27B) is a common autosomal dominant ataxia caused by an intronic GAA•TTC repeat expansion in FGF14. Neuropathological studies have shown that neuronal loss is largely restricted to the cerebellum. Although the repeat locus is highly unstable during intergenerational transmission, it remains unknown whether it exhibits cerebral mosaicism and progressive instability throughout life. We conducted an analysis of the FGF14 GAA•TTC repeat somatic instability across 156 serial blood samples from 69 individuals, fibroblasts, induced pluripotent stem cells and post-mortem brain tissues from six controls and six patients with SCA27B, alongside methylation profiling using targeted long-read sequencing. Peripheral tissues exhibited minimal somatic instability, which did not significantly change over periods of more than 20 years. In post-mortem brains, the GAA•TTC repeat was remarkably stable across all regions, except in the cerebellar hemispheres and vermis. The levels of somatic expansion in the cerebellar hemispheres and vermis were, on average, 3.15 and 2.72 times greater relative to other examined brain regions, respectively. Additionally, levels of somatic expansion in the brain increased with repeat length and tissue expression of FGF14. We found no significant difference in methylation of wild-type and expanded FGF14 alleles in post-mortem cerebellar hemispheres between patients and controls. In conclusion, our study revealed that the FGF14 GAA•TTC repeat exhibits a cerebellar-specific expansion bias, which may explain the pure cerebellar involvement in SCA27B.
Keywords: GAA-FGF14 ataxia; SCA27B; mosaicism; repeat expansion; repeat expansion disorder; spinocerebellar ataxia 27B.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
I.W.D. manages a fee-for-service sequencing facility at the Garvan Institute of Medical Research that is a customer of Oxford Nanopore Technologies but has no further financial relationship. I.W.D. has previously received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences. M.N. received consultancy honoraria from Reata Pharmaceuticals unrelated to this work. L.M.P. received consultancy honoraria from Biogen unrelated to this work. S.Z. has received consultancy honoraria from Neurogene, Aeglea BioTherapeutics, Applied Therapeutics, and is an unpaid officer of the TGP foundation, all unrelated to the present manuscript. A.D. serves as an advisor at Critical Path Ataxia Therapeutics Consortium and her Institution (Paris Brain Institute) receives consulting fees on her behalf from Biogen, Huntix, UCB, as well as research grants from the NIH, ANR and holds partly a Patent B 06291873.5 on ‘Anaplerotic therapy of Huntington disease and other polyglutamine diseases’. All other authors report no competing interests.
Figures

Update of
-
Somatic instability of the FGF14 -SCA27B GAA•TTC repeat reveals a marked expansion bias in the cerebellum.medRxiv [Preprint]. 2024 Jul 3:2024.07.01.24309777. doi: 10.1101/2024.07.01.24309777. medRxiv. 2024. Update in: Brain. 2025 Apr 03;148(4):1258-1270. doi: 10.1093/brain/awae312. PMID: 39006414 Free PMC article. Updated. Preprint.
References
-
- Wilke C, Pellerin D, Mengel D, et al. GAA-FGF14 ataxia (SCA27B): Phenotypic profile, natural history progression and 4-aminopyridine treatment response. Brain. 2023;146:4144–4157. - PubMed
MeSH terms
Substances
Grants and funding
- MRC_/Medical Research Council/United Kingdom
- 2R01NS072248-11A1/NIH National Institutes of Neurological Disorders and Stroke
- 13338/Fondation pour la Recherche Médicale
- DP1DA056018/DA/NIDA NIH HHS/United States
- MSA Trust
- HHSN261201500003/CA/NCI NIH HHS/United States
- NS081366/National Institutes of Neurological Disorders and Stroke
- Manx MSA Trust
- 189963/CAPMC/ CIHR/Canada
- National Health and Medical Research Council
- Michael J Fox Foundation
- MSA Coalition
- Alzheimer's Research UK
- R01 NS072248/NS/NINDS NIH HHS/United States
- DP1 DA056018/DA/NIDA NIH HHS/United States
- UCLH Biomedical Research Centre
- Fidelity Trust
- King Baudouin Foundation
- Fondation Groupe Monaco
- Friedreich's Ataxia Research Alliance
- 565908/Guarantors of Brain
- NH/NIH HHS/United States
- 1173594/Medical Research Future Fund
- National Institute for Health and Care Research
- Ataxia UK
- WT_/Wellcome Trust/United Kingdom
- 540868/Association of British Neurologists
- R01 NS081366/NS/NINDS NIH HHS/United States
- Rosetrees Trust
- UCLH/UCL Biomedical Research Centre
LinkOut - more resources
Full Text Sources
Miscellaneous

