Evidence for a hydrogen sulfide-sensing E3 ligase in yeast
- PMID: 39378345
- PMCID: PMC11538405
- DOI: 10.1093/genetics/iyae154
Evidence for a hydrogen sulfide-sensing E3 ligase in yeast
Abstract
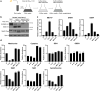
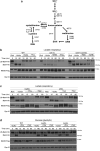
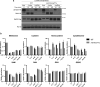
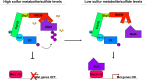
In yeast, control of sulfur amino acid metabolism relies upon Met4, a transcription factor that activates the expression of a network of enzymes responsible for the biosynthesis of cysteine and methionine. In times of sulfur abundance, the activity of Met4 is repressed via ubiquitination by the SCFMet30 E3 ubiquitin ligase, but the mechanism by which the F-box protein Met30 senses sulfur status to tune its E3 ligase activity remains unresolved. Herein, we show that Met30 responds to flux through the trans-sulfuration pathway to regulate the MET gene transcriptional program. In particular, Met30 is responsive to the biological gas hydrogen sulfide, which is sufficient to induce ubiquitination of Met4 in vivo. Additionally, we identify important cysteine residues in Met30's WD-40 repeat region that sense the availability of sulfur in the cell. Our findings reveal how SCFMet30 dynamically senses the flow of sulfur metabolites through the trans-sulfuration pathway to regulate the synthesis of these special amino acids.
Keywords: E3 ubiquitin ligase; amino acid; metabolism; nutrients; sensor; sulfur; yeast.
© The Author(s) 2024. Published by Oxford University Press on behalf of The Genetics Society of America.
Conflict of interest statement
Conflicts of interest The authors declare no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

