Type I IFN drives unconventional IL-1β secretion in lupus monocytes
- PMID: 39378884
- PMCID: PMC11563874
- DOI: 10.1016/j.immuni.2024.09.004
Type I IFN drives unconventional IL-1β secretion in lupus monocytes
Abstract
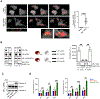
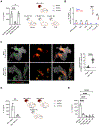
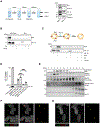
Opsonization of red blood cells that retain mitochondria (Mito+ RBCs), a feature of systemic lupus erythematosus (SLE), triggers type I interferon (IFN) production in macrophages. We report that monocytes (Mos) co-produce IFN and mature interleukin-1β (mIL-1β) upon Mito+ RBC opsonization. IFN expression depended on cyclic GMP-AMP synthase (cGAS) and RIG-I-like receptors' (RLRs) sensing of Mito+ RBC-derived mitochondrial DNA (mtDNA) and mtRNA, respectively. Interleukin-1β (IL-1β) production was initiated by the RLR antiviral signaling adaptor (MAVS) pathway recognition of Mito+ RBC-derived mtRNA. This led to the cytosolic release of Mo mtDNA, which activated the inflammasome. Importantly, mIL-1β secretion was independent of gasdermin D (GSDMD) and pyroptosis but relied on IFN-inducible myxovirus-resistant protein 1 (MxA), which facilitated the incorporation of mIL-1β into a trans-Golgi network (TGN)-mediated secretory pathway. RBC internalization identified a subset of blood Mo expressing IFN-stimulated genes (ISGs) that released mIL-1β and expanded in SLE patients with active disease.
Keywords: MxA; NLRP3; inflammasome; monocytes; red blood cells; systemic lupus erythematosus; type I interferon.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests V.P. has received consulting honoraria from Sanofi, Astra Zeneca, Regeneron, and Moderna and is the recipient of a research grant from Sanofi and a contract from Astra Zeneca. J.F.B. is an employee of Immunoledge LLC. V.P. and J.F.B. receive royalties for use of Canakinumab in sJIA.
Figures




Comment in
-
Mx1-ing it up-Mitochondrial relay for interferon-dependent, unconventional IL-1β release in SLE monocytes.Immunity. 2024 Nov 12;57(11):2483-2486. doi: 10.1016/j.immuni.2024.10.005. Immunity. 2024. PMID: 39536711 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

