Coronavirus envelope protein activates TMED10-mediated unconventional secretion of inflammatory factors
- PMID: 39379362
- PMCID: PMC11461611
- DOI: 10.1038/s41467-024-52818-0
Coronavirus envelope protein activates TMED10-mediated unconventional secretion of inflammatory factors
Abstract
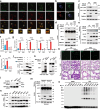
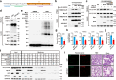
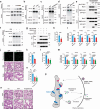
The precise cellular mechanisms underlying heightened proinflammatory cytokine production during coronavirus infection remain incompletely understood. Here we identify the envelope (E) protein in severe coronaviruses (SARS-CoV-2, SARS, or MERS) as a potent inducer of interleukin-1 release, intensifying lung inflammation through the activation of TMED10-mediated unconventional protein secretion (UcPS). In contrast, the E protein of mild coronaviruses (229E, HKU1, or OC43) demonstrates a less pronounced effect. The E protein of severe coronaviruses contains an SS/DS motif, which is not present in milder strains and facilitates interaction with TMED10. This interaction enhances TMED10-oligomerization, facilitating UcPS cargo translocation into the ER-Golgi intermediate compartment (ERGIC)-a pivotal step in interleukin-1 UcPS. Progesterone analogues were identified as compounds inhibiting E-enhanced release of proinflammatory factors and lung inflammation in a Mouse Hepatitis Virus (MHV) infection model. These findings elucidate a molecular mechanism driving coronavirus-induced hyperinflammation, proposing the E-TMED10 interaction as a potential therapeutic target to counteract the adverse effects of coronavirus-induced inflammation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 32370728/National Natural Science Foundation of China (National Science Foundation of China)
- 32200553/National Natural Science Foundation of China (National Science Foundation of China)
- 92254302/National Natural Science Foundation of China (National Science Foundation of China)
- 32225013/National Natural Science Foundation of China (National Science Foundation of China)
- 32130023/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Miscellaneous

