Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery
- PMID: 39379370
- PMCID: PMC11461821
- DOI: 10.1038/s41467-024-52950-x
Structure of a Rhs effector clade domain provides mechanistic insights into type VI secretion system toxin delivery
Abstract
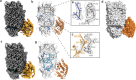
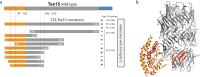
The type VI secretion system (T6SS) is a molecular machine utilised by many Gram-negative bacteria to deliver antibacterial toxins into adjacent cells. Here we present the structure of Tse15, a T6SS Rhs effector from the nosocomial pathogen Acinetobacter baumannii. Tse15 forms a triple layered β-cocoon Rhs domain with an N-terminal α-helical clade domain and an unfolded C-terminal toxin domain inside the Rhs cage. Tse15 is cleaved into three domains, through independent auto-cleavage events involving aspartyl protease activity for toxin self-cleavage and a nucleophilic glutamic acid for N-terminal clade cleavage. Proteomic analyses identified that significantly more peptides from the N-terminal clade and toxin domains were secreted than from the Rhs cage, suggesting toxin delivery often occurs without the cage. We propose the clade domain acts as an internal chaperone to mediate toxin tethering to the T6SS machinery. Conservation of the clade domain in other Gram-negative bacteria suggests this may be a common mechanism for delivery.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures


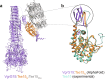


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- 1128981/Department of Health | National Health and Medical Research Council (NHMRC)
- 1165036/Department of Health | National Health and Medical Research Council (NHMRC)
- 1165036/Department of Health | National Health and Medical Research Council (NHMRC)
- 1128981/Department of Health | National Health and Medical Research Council (NHMRC)
- 1165036/Department of Health | National Health and Medical Research Council (NHMRC)
LinkOut - more resources
Full Text Sources

