DELE1 maintains muscle proteostasis to promote growth and survival in mitochondrial myopathy
- PMID: 39379554
- PMCID: PMC11574132
- DOI: 10.1038/s44318-024-00242-x
DELE1 maintains muscle proteostasis to promote growth and survival in mitochondrial myopathy
Abstract
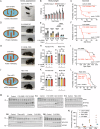
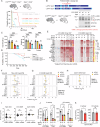
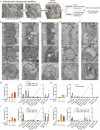
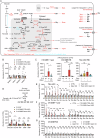
Mitochondrial dysfunction causes devastating disorders, including mitochondrial myopathy, but how muscle senses and adapts to mitochondrial dysfunction is not well understood. Here, we used diverse mouse models of mitochondrial myopathy to show that the signal for mitochondrial dysfunction originates within mitochondria. The mitochondrial proteins OMA1 and DELE1 sensed disruption of the inner mitochondrial membrane and, in response, activated the mitochondrial integrated stress response (mt-ISR) to increase the building blocks for protein synthesis. In the absence of the mt-ISR, protein synthesis in muscle was dysregulated causing protein misfolding, and mice with early-onset mitochondrial myopathy failed to grow and survive. The mt-ISR was similar following disruptions in mtDNA maintenance (Tfam knockout) and mitochondrial protein misfolding (CHCHD10 G58R and S59L knockin) but heterogenous among mitochondria-rich tissues, with broad gene expression changes observed in heart and skeletal muscle and limited changes observed in liver and brown adipose tissue. Taken together, our findings identify that the DELE1 mt-ISR mediates a similar response to diverse forms of mitochondrial stress and is critical for maintaining growth and survival in early-onset mitochondrial myopathy.
Keywords: Mitochondria Unfolded Protein Response (mt-UPR); Mitochondrial Disorders; Mitohormesis; Mitonuclear Communication; Mitophagy.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Figures






Update of
-
DELE1 promotes translation-associated homeostasis, growth, and survival in mitochondrial myopathy.bioRxiv [Preprint]. 2024 Feb 29:2024.02.29.582673. doi: 10.1101/2024.02.29.582673. bioRxiv. 2024. Update in: EMBO J. 2024 Nov;43(22):5548-5585. doi: 10.1038/s44318-024-00242-x. PMID: 38529505 Free PMC article. Updated. Preprint.
References
-
- Ahola S, Rivera Mejías P, Hermans S, Chandragiri S, Giavalisco P, Nolte H, Langer T (2022) OMA1-mediated integrated stress response protects against ferroptosis in mitochondrial cardiomyopathy. Cell Metab 34:1875–1891.e7 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

