GWAS of multiple neuropathology endophenotypes identifies new risk loci and provides insights into the genetic risk of dementia
- PMID: 39379761
- PMCID: PMC11549054
- DOI: 10.1038/s41588-024-01939-9
GWAS of multiple neuropathology endophenotypes identifies new risk loci and provides insights into the genetic risk of dementia
Erratum in
-
Author Correction: GWAS of multiple neuropathology endophenotypes identifies new risk loci and provides insights into the genetic risk of dementia.Nat Genet. 2025 Jul;57(7):1788. doi: 10.1038/s41588-024-02045-6. Nat Genet. 2025. PMID: 40527984 Free PMC article. No abstract available.
-
Publisher Correction: GWAS of multiple neuropathology endophenotypes identifies new risk loci and provides insights into the genetic risk of dementia.Nat Genet. 2025 Jul;57(7):1791. doi: 10.1038/s41588-024-02046-5. Nat Genet. 2025. PMID: 40527985 Free PMC article. No abstract available.
Abstract
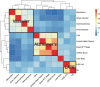
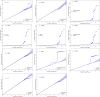
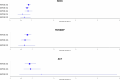
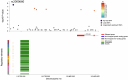
Genome-wide association studies (GWAS) have identified >80 Alzheimer's disease and related dementias (ADRD)-associated genetic loci. However, the clinical outcomes used in most previous studies belie the complex nature of underlying neuropathologies. Here we performed GWAS on 11 ADRD-related neuropathology endophenotypes with participants drawn from the following three sources: the National Alzheimer's Coordinating Center, the Religious Orders Study and Rush Memory and Aging Project, and the Adult Changes in Thought study (n = 7,804 total autopsied participants). We identified eight independent significantly associated loci, of which four were new (COL4A1, PIK3R5, LZTS1 and APOC2). Separately testing known ADRD loci, 19 loci were significantly associated with at least one neuropathology after false-discovery rate adjustment. Genetic colocalization analyses identified pleiotropic effects and quantitative trait loci. Methylation in the cerebral cortex at two sites near APOC2 was associated with cerebral amyloid angiopathy. Studies that include neuropathology endophenotypes are an important step in understanding the mechanisms underlying genetic ADRD risk.
© 2024. The Author(s).
Conflict of interest statement
J.A.S. reported personal fees from the Observational Study Monitoring Board Framingham, Observational Study Monitoring Board Discovery (National Institute of Neurological Disorders and Stroke) and Takeda Pharma. A.J.S. reported support from Avid Radiopharmaceuticals, a subsidiary of Eli Lilly (in kind contribution of positron emission tomography tracer precursor), and participated in Scientific Advisory Boards (Bayer Oncology, Eisai, Novo Nordisk and Siemens Medical Solutions) and an Observational Study Monitoring Board (MESA, NIH NHLBI), as well as several other NIA External Advisory Committees. He also serves as editor-in-chief of Brain Imaging and Behavior, a Springer Nature Journal. He was not involved in the editorial handling of this Nature Genetics paper (journals within the Springer Nature Portfolio are editorially independent). The other authors declare no competing interests.
Figures







References
MeSH terms
Grants and funding
- R01 AG043379/AG/NIA NIH HHS/United States
- P50 AG047266/AG/NIA NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- R01 AG034374/AG/NIA NIH HHS/United States
- R35 GM138636/GM/NIGMS NIH HHS/United States
- P50 AG033514/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- R01 AG047976/AG/NIA NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- P30 AG053760/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- R56 AG057191/AG/NIA NIH HHS/United States
- P50 AG023501/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- P30 AG010124/AG/NIA NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- U01 AG058654/AG/NIA NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- RC2 AG036528/AG/NIA NIH HHS/United States
- R01 AG042210/AG/NIA NIH HHS/United States
- UH2 NS100599/NS/NINDS NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- R01 AG061111/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- P01 AG078116/AG/NIA NIH HHS/United States
- U01 AG072177/AG/NIA NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- U01 NS100599/NS/NINDS NIH HHS/United States
- R01 NS078009/NS/NINDS NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- U19 AG074879/AG/NIA NIH HHS/United States
- R01 AG056352/AG/NIA NIH HHS/United States
- P30 AG035982/AG/NIA NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- RF1 NS118584/NS/NINDS NIH HHS/United States
- U01 AG068057/AG/NIA NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- P30 AG008051/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- P50 AG047270/AG/NIA NIH HHS/United States
- P30 AG072980/AG/NIA NIH HHS/United States
- K25 AG055620/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- R01 AG033678/AG/NIA NIH HHS/United States
- P30 AG049638/AG/NIA NIH HHS/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- RF1 AG054057/AG/NIA NIH HHS/United States
- U19 AG024904/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- U24 AG041689/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P30 AG008017/AG/NIA NIH HHS/United States
- R01 AG036042/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- RF1AG082339/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- P50 AG025688/AG/NIA NIH HHS/United States
- F30 NS124136/NS/NINDS NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- R01 AG022018/AG/NIA NIH HHS/United States
- R01 AG068331/AG/NIA NIH HHS/United States
- P50 AG047366/AG/NIA NIH HHS/United States
- R01 LM012535/LM/NLM NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- RF1 AG082339/AG/NIA NIH HHS/United States
- R01 AG040039/AG/NIA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- P30 AG028383/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- R01 AG082730/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

