Prediction of homologous recombination deficiency from routine histology with attention-based multiple instance learning in nine different tumor types
- PMID: 39379982
- PMCID: PMC11462727
- DOI: 10.1186/s12915-024-02022-9
Prediction of homologous recombination deficiency from routine histology with attention-based multiple instance learning in nine different tumor types
Abstract
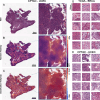
Background: Homologous recombination deficiency (HRD) is recognized as a pan-cancer predictive biomarker that potentially indicates who could benefit from treatment with PARP inhibitors (PARPi). Despite its clinical significance, HRD testing is highly complex. Here, we investigated in a proof-of-concept study whether Deep Learning (DL) can predict HRD status solely based on routine hematoxylin & eosin (H&E) histology images across nine different cancer types.
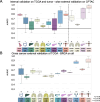
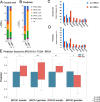
Methods: We developed a deep learning pipeline with attention-weighted multiple instance learning (attMIL) to predict HRD status from histology images. As part of our approach, we calculated a genomic scar HRD score by combining loss of heterozygosity (LOH), telomeric allelic imbalance (TAI), and large-scale state transitions (LST) from whole genome sequencing (WGS) data of n = 5209 patients across two independent cohorts. The model's effectiveness was evaluated using the area under the receiver operating characteristic curve (AUROC), focusing on its accuracy in predicting genomic HRD against a clinically recognized cutoff value.
Results: Our study demonstrated the predictability of genomic HRD status in endometrial, pancreatic, and lung cancers reaching cross-validated AUROCs of 0.79, 0.58, and 0.66, respectively. These predictions generalized well to an external cohort, with AUROCs of 0.93, 0.81, and 0.73. Moreover, a breast cancer-trained image-based HRD classifier yielded an AUROC of 0.78 in the internal validation cohort and was able to predict HRD in endometrial, prostate, and pancreatic cancer with AUROCs of 0.87, 0.84, and 0.67, indicating that a shared HRD-like phenotype occurs across these tumor entities.
Conclusions: This study establishes that HRD can be directly predicted from H&E slides using attMIL, demonstrating its applicability across nine different tumor types.
Keywords: Artificial intelligence; DNA repair mechanism; Deep learning; Homologous recombination deficiency; Mpathology; Pan-cancer study.
© 2024. The Author(s).
Conflict of interest statement
JNK reports consulting services for Owkin, France, Panakeia, UK, and DoMore Diagnostics, Norway, and has received honoraria for lectures by MSD, Eisai, and Fresenius. JSRF reports a leadership (board of directors) role at Grupo Oncoclinicas, stock or other ownership interests at Repare Therapeutics and Paige.AI, and a consulting or Advisory Role at Genentech/Roche, Invicro, Ventana Medical Systems, Volition RX, Paige.AI, Goldman Sachs, Bain Capital, Novartis, Repare Therapeutics, Lilly, Saga Diagnostics, Swarm and Personalis. No other potential conflicts of interest are reported by any of the authors.
Figures

Update of
-
Direct prediction of Homologous Recombination Deficiency from routine histology in ten different tumor types with attention-based Multiple Instance Learning: a development and validation study.medRxiv [Preprint]. 2023 Mar 10:2023.03.08.23286975. doi: 10.1101/2023.03.08.23286975. medRxiv. 2023. Update in: BMC Biol. 2024 Oct 8;22(1):225. doi: 10.1186/s12915-024-02022-9. PMID: 36945540 Free PMC article. Updated. Preprint.
References
-
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

