Heparin-enriched plasma proteome is significantly altered in Alzheimer's disease
- PMID: 39380021
- PMCID: PMC11460197
- DOI: 10.1186/s13024-024-00757-1
Heparin-enriched plasma proteome is significantly altered in Alzheimer's disease
Abstract
Introduction: Heparin binding proteins (HBPs) with roles in extracellular matrix assembly are strongly correlated to β-amyloid (Aβ) and tau pathology in Alzheimer's disease (AD) brain and cerebrospinal fluid (CSF). However, it remains challenging to detect these proteins in plasma using standard mass spectrometry-based proteomic approaches.
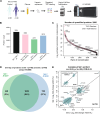
Methods: We employed heparin-affinity chromatography, followed by off-line fractionation and tandem mass tag mass spectrometry (TMT-MS), to enrich HBPs from plasma obtained from AD (n = 62) and control (n = 47) samples. These profiles were then correlated to Aβ, tau and phosphorylated tau (pTau) CSF biomarkers and plasma pTau181 from the same individuals, as well as a consensus brain proteome network to assess the overlap with AD brain pathophysiology.
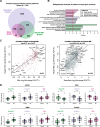
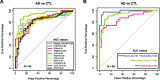
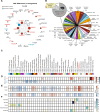
Results: Heparin enrichment from plasma was highly reproducible, enriched well-known HBPs like APOE and thrombin, and depleted high-abundant proteins such as albumin. A total of 2865 proteins, spanning 10 orders of magnitude in abundance, were measured across 109 samples. Compared to the consensus AD brain protein co-expression network, we observed that specific plasma proteins exhibited consistent direction of change in both brain and plasma, whereas others displayed divergent changes, highlighting the complex interplay between the two compartments. Elevated proteins in AD plasma, when compared to controls, included members of the matrisome module in brain that accumulate with Aβ deposits, such as SMOC1, SMOC2, SPON1, MDK, OLFML3, FRZB, GPNMB, and the APOE4 proteoform. Additionally, heparin-enriched proteins in plasma demonstrated significant correlations with conventional AD CSF biomarkers, including Aβ, total tau, pTau, and plasma pTau181. A panel of five plasma proteins classified AD from control individuals with an area under the curve (AUC) of 0.85. When combined with plasma pTau181, the panel significantly improved the classification performance of pTau181 alone, increasing the AUC from 0.93 to 0.98. This suggests that the heparin-enriched plasma proteome captures additional variance in cognitive dementia beyond what is explained by pTau181.
Conclusion: These findings support the utility of a heparin-affinity approach coupled with TMT-MS for enriching amyloid-associated proteins, as well as a wide spectrum of plasma biomarkers that reflect pathological changes in the AD brain.
Keywords: Alzheimer’s disease; Amyloid; Biomarkers; Cerebrospinal fluid; Heparan sulfate proteoglycans; Heparin; Plasma; Proteomics.
© 2024. The Author(s).
Conflict of interest statement
A.I.L, N.T.S. and D.M.D. are co-founders of Emtherapro Inc.
Figures




Update of
-
Heparin-enriched plasma proteome is significantly altered in Alzheimer's Disease.Res Sq [Preprint]. 2024 Feb 23:rs.3.rs-3933136. doi: 10.21203/rs.3.rs-3933136/v1. Res Sq. 2024. Update in: Mol Neurodegener. 2024 Oct 8;19(1):67. doi: 10.1186/s13024-024-00757-1. PMID: 38464223 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

