Testicular large B-cell lymphoma is genetically similar to PCNSL and distinct from nodal DLBCL
- PMID: 39380845
- PMCID: PMC11456803
- DOI: 10.1002/hem3.70024
Testicular large B-cell lymphoma is genetically similar to PCNSL and distinct from nodal DLBCL
Abstract
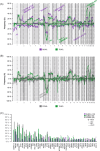
Testicular large B-cell lymphoma (TLBCL) is an infrequent and aggressive lymphoma arising in an immune-privileged site and has recently been recognized as a distinct entity from diffuse large B-cell lymphoma (DLBCL). We describe the genetic features of TLBCL and compare them with published series of nodal DLBCL and primary large B-cell lymphomas of the CNS (PCNSL). We collected 61 patients with TLBCL. We performed targeted next-generation sequencing, copy number arrays, and fluorescent in situ hybridization to assess chromosomal rearrangements in 40 cases with available material. Seventy percent of the cases showed localized stages. BCL6 rearrangements were detected in 36% of cases, and no concomitant BCL2 and MYC rearrangements were found. TLBCL had fewer copy number alterations (p < 0.04) but more somatic variants (p < 0.02) than nodal DLBCL and had more frequent 18q21.32-q23 (BCL2) gains and 6q and 9p21.3 (CDKN2A/B) deletions. PIM1, MYD88 L265P , CD79B, TBL1XR1, MEF2B, CIITA, EP300, and ETV6 mutations were more frequent in TLBCL, and BCL10 mutations in nodal DLBCL. There were no major genetic differences between TLBCL and PCNSL. Localized or disseminated TLBCL displayed similar genomic profiles. Using LymphGen, the majority of cases were classified as MCD. However, we observed a subgroup of patients classified as BN2, both in localized and disseminated TLBCL, suggesting a degree of genetic heterogeneity in the TLBCL genetic profile. TLBCL has a distinctive genetic profile similar to PCNSL, supporting its recognition as a separate entity from DLBCL and might provide information to devise targeted therapeutic approaches.
© 2024 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
Ferran Nadeu has received honoraria from Janssen, AbbVie, AstraZeneca, and SOPHiA GENETICS for speaking at educational activities; has received research support from Gilead; and has licensed the use of the protected IgCaller algorithm to Diagnóstica Longwood. Tycho Baumann has received consulting fees or honoraria from Janssen, Roche, Novartis, Merck, Gilead/Kite, Incyte, Lilly, Abbvie, AstraZeneca, and BeiGene. Alejandro Martin García‐Sancho has received consulting fees or honoraria from Janssen, Roche, BMS/Celgene, Kyowa Kirin, Clinigen, EUSAPharma, Novartis, Gilead/Kite, Incyte, Lilly, Takeda, ADC Therapeutics America, Miltenyi, Ideogen, Abbvie, and BeiGene. Elías Campo has been a consultant for GenMab, and Takeda; has received research support from AstraZeneca; received honoraria from Janssen, EUSPharma, Takeda, and Roche for speaking at educational activities; and is an inventor on a Lymphoma and Leukemia Molecular Profiling Project patent “Method for subtyping lymphoma subtypes by means of expression profiling” (PCT/US2014/64161) and a bioinformatic tool (IgCaller) licensed to Diagnostic Longwood. Eva Giné has received honoraria or consulting fees from Gilead, Kite Pharma, Janssen, Genmab, Miltenyi, and Lilly; has received research support from Janssen and travel expenses from Gilead and Kite Pharma. Armando López Guillermo served on the advisory board of Roche, Celgene, Novartis, and Gilead/Kite, received grants from Celgene and Gilead/Kite, and travel expenses from Kite/Gilead. The remaining authors declare no competing interests.
Figures


References
-
- Cheah CY, Wirth A, Seymour JF. Primary testicular lymphoma. Blood. 2014;123:486‐493. - PubMed
-
- Møller MB, d'Amore F, Christensen BE. Testicular lymphoma: a population‐based study of incidence, clinicopathological correlations and prognosis. Eur J Cancer. 1994;30A:1760‐1764. - PubMed
-
- Menter T, Ernst M, Drachneris J, et al. Phenotype profiling of primary testicular diffuse large B‐cell lymphomas. Hematol Oncol. 2014;32:72‐81. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
