Co-regulation of innate and adaptive immune responses induced by ID93+GLA-SE vaccination in humans
- PMID: 39381003
- PMCID: PMC11458388
- DOI: 10.3389/fimmu.2024.1441944
Co-regulation of innate and adaptive immune responses induced by ID93+GLA-SE vaccination in humans
Abstract
Introduction: Development of an effective vaccine against tuberculosis is a critical step towards reducing the global burden of disease. A therapeutic vaccine might also reduce the high rate of TB recurrence and help address the challenges of drug-resistant strains. ID93+GLA-SE is a candidate subunit vaccine that will soon be evaluated in a phase 2b efficacy trial for prevention of recurrent TB among patients undergoing TB treatment. ID93+GLA-SE vaccination was shown to elicit robust CD4+ T cell and IgG antibody responses among recently treated TB patients in the TBVPX-203 Phase 2a study (NCT02465216), but the mechanisms underlying these responses are not well understood.
Methods: In this study we used specimens from TBVPX-203 participants to describe the changes in peripheral blood gene expression that occur after ID93+GLA-SE vaccination.
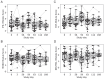
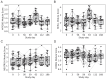
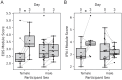
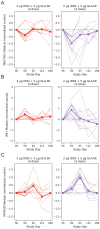
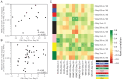
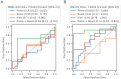
Results: Analyses revealed several distinct modules of co-varying genes that were either up- or down-regulated after vaccination, including genes associated with innate immune pathways at 3 days post-vaccination and genes associated with lymphocyte expansion and B cell activation at 7 days post-vaccination. Notably, the regulation of these gene modules was affected by the dose schedule and by participant sex, and early innate gene signatures were correlated with the ID93-specific CD4+ T cell response.
Discussion: The results provide insight into the complex interplay of the innate and adaptive arms of the immune system in developing responses to vaccination with ID93+GLA-SE and demonstrate how dosing and schedule can affect vaccine responses.
Keywords: RNA sequencing; innate; systems immunology; tuberculosis; vaccines.
Copyright © 2024 Fiore-Gartland, Srivastava, Seese, Day, Penn-Nicholson, Luabeya, Du Plessis, Loxton, Bekker, Diacon, Walzl, Sagawa, Reed, Scriba, Hatherill and Coler.
Conflict of interest statement
Author SR was employed by the company HDT Bio Corporation. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



References
-
- World Health Organization . Global Tuberculosis Report 2023 . Available online at: https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... (Accessed May 4, 2024).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

