Fast Context-Aware Analysis of Genome Annotation Colocalization
- PMID: 39381845
- PMCID: PMC11698669
- DOI: 10.1089/cmb.2024.0667
Fast Context-Aware Analysis of Genome Annotation Colocalization
Abstract
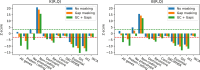
An annotation is a set of genomic intervals sharing a particular function or property. Examples include genes or their exons, sequence repeats, regions with a particular epigenetic state, and copy number variants. A common task is to compare two annotations to determine if one is enriched or depleted in the regions covered by the other. We study the problem of assigning statistical significance to such a comparison based on a null model representing random unrelated annotations. To incorporate more background information into such analyses, we propose a new null model based on a Markov chain that differentiates among several genomic contexts. These contexts can capture various confounding factors, such as GC content or assembly gaps. We then develop a new algorithm for estimating p-values by computing the exact expectation and variance of the test statistic and then estimating the p-value using a normal approximation. Compared to the previous algorithm by Gafurov et al., the new algorithm provides three advances: (1) the running time is improved from quadratic to linear or quasi-linear, (2) the algorithm can handle two different test statistics, and (3) the algorithm can handle both simple and context-dependent Markov chain null models. We demonstrate the efficiency and accuracy of our algorithm on synthetic and real data sets, including the recent human telomere-to-telomere assembly. In particular, our algorithm computed p-values for 450 pairs of human genome annotations using 24 threads in under three hours. Moreover, the use of genomic contexts to correct for GC bias resulted in the reversal of some previously published findings.
Keywords: Markov chains; colocalization; genome annotation.
Figures







References
-
- Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol 1997;268(1):78–94. - PubMed
-
- Durbin R, Eddy SR, Krogh A, et al. Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids. Cambridge University Press; 1998.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

