SRSF1 Is Crucial for Maintaining Satellite Cell Homeostasis During Skeletal Muscle Growth and Regeneration
- PMID: 39381961
- PMCID: PMC11634495
- DOI: 10.1002/jcsm.13607
SRSF1 Is Crucial for Maintaining Satellite Cell Homeostasis During Skeletal Muscle Growth and Regeneration
Abstract
Background: The splicing factor SRSF1 emerges as a mater regulator of cell proliferation, displaying high expression in actively proliferative satellite cells (SCs). In SRSF1 knockout mice (KO) generated via MyoD-Cre, early mortality and muscle atrophy are observed during postnatal muscle growth. Despite these findings, the precise mechanisms through which SRSF1 loss influences SCs' functions and its role in muscle regeneration remain to be elucidated.
Methods: To unravel the exact mechanisms underlying the impact of SRSF1 deficiency SC functions, we employed single-cell RNA sequencing (scRNA-seq) on a mononuclear cell suspension isolated from the newborn diaphragm of KO and control mice. Concurrently, we subjected diaphragm muscles to RNA-seq analysis to identify dysregulated splicing events associated with SRSF1 deletion. For the analysis of the effect of SRSF1 deletion on muscle regeneration, we generated mice with inducible SC-specific Srsf1 ablation through Pax7-CreER. SRSF1 ablation was induced by intraperitoneal injection of tamoxifen. Using cardiotoxin-induced muscle injury, we examined the consequences of SRSF1 depletion on SC function through HE staining, immunostaining and EdU incorporation assay. C2C12 myoblasts and isolated myoblasts were employed to assess stem cell function and senescence.
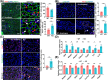
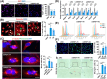
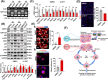
Results: Utilizing scRNA-seq analysis, we observed a noteworthy increase in activated and proliferating myoblasts when SRSF1 was absent. This increase was substantial, with the proportion rising from 28.68% in the control group to 77.06% in the knockout group. However, these myoblasts experienced mitotic abnormalities in the absence of SRSF1, resulting in cell cycle arrest and the onset of cellular senescence. In the knockout mice, the proportion of Pax7+ cells within improper niche positioning increased significantly to 25% compared to 12% in the control cells (n ≥ 10, p < 0.001). Furthermore, there was an observation of persistent cell cycle exit specifically in the Pax7+ cells deficient in SRSF1 (n = 6, p < 0.001). SRSF1 plays a pivotal role in regulating the splicing of Fgfr1op2, favouring the full-length isoform crucial for mitotic spindle organization. Disrupting SRSF1 in C2C12 and primary myoblasts results in multipolar spindle formation (p < 0.001) and dysregulated splicing of Fgfr1op2 and triggers cellular senescence. Consequently, adult SCs lacking SRSF1 initially activate upon injury but face substantial challenge in proliferation (n = 4, p < 0.001), leading to a failure in muscle regeneration.
Conclusions: SRSF1 plays a critical role in SCs by ensuring proper splicing, maintaining mitotic progression and preventing premature senescence. These findings underscore the significant role of SRSF1 in controlling SC proliferation during skeletal muscle growth and regeneration.
Keywords: SRSF1; cellular senescence; dysregulated splicing; muscle regeneration; satellite cells; scRNA‐seq.
© 2024 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Sousa‐Victor P., Garcia‐Prat L., and Munoz‐Canoves P., “Control of Satellite Cell Function in Muscle Regeneration and Its Disruption in Ageing,” Nature Reviews. Molecular Cell Biology 23 (2022): 204–226. - PubMed
-
- Fukada S., Uezumi A., Ikemoto M., et al., “Molecular Signature of Quiescent Satellite Cells in Adult Skeletal Muscle,” Stem Cells 25 (2007): 2448–2459. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

