Maternal sleep deprivation disrupts glutamate metabolism in offspring rats
- PMID: 39382081
- PMCID: PMC11668958
- DOI: 10.24272/j.issn.2095-8137.2024.250
Maternal sleep deprivation disrupts glutamate metabolism in offspring rats
Abstract
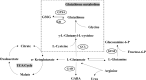
Maternal sleep deprivation (MSD) has emerged as a significant public health concern, yet its effects on offspring metabolism remain poorly understood. This study investigated the metabolomic implications of MSD on offspring cognitive development, with a particular focus on alterations in glutamate metabolism. Pregnant rats were subjected to sleep deprivation during late gestation. Plasma and brain samples from their offspring were collected at different postnatal days (P1, P7, P14, and P56) and analyzed using untargeted metabolomics with liquid chromatography-mass spectrometry. Metabolomic analysis revealed significant differences in various amino acids, including L-glutamate, L-phenylalanine, L-tyrosine, and L-tryptophan, which are crucial for cognitive function. Subsequent differential analysis and partial least squares discriminant analysis (sPLS-DA) demonstrated a gradual reduction in these metabolic differences in the brain as the offspring underwent growth and development. KEGG pathway analysis revealed differential regulation of several pathways, including alanine, aspartate, and glutamate metabolism, glutathione metabolism, arginine biosynthesis, aminoacyl-tRNA biosynthesis, histidine metabolism, and taurine and hypotaurine metabolism, at different developmental stages. Mantel and Spearman analyses indicated that the observed changes in metabolites in MSD progeny may be related to various gut microbes, Ruminococcus_1, Ruminococcaceae_UCG-005, and Eubacterium_coprostanoligenes_group. Biochemical assays further demonstrated developmental changes in the L-glutamate metabolic pathway. Collectively, these findings suggest that MSD not only affects maternal well-being but also has enduring metabolic consequences for offspring, particularly impacting pathways linked to cognitive function. This highlights the importance of addressing maternal sleep health to mitigate potential long-term consequences for offspring.
母体睡眠剥夺(MSD)已成为一个重要的社会问题,但其对后代代谢的影响仍知之甚少。该研究探讨了MSD对后代认知发育的代谢组学影响,特别是对谷氨酸代谢的改变。实验中,对妊娠晚期的大鼠进行睡眠剥夺处理,并在不同生后日(P1、P7、P14和P56)采集其子代的血浆和脑组织样本,通过液相色谱-质谱联用的非靶向代谢组学分析进行研究。代谢组学分析显示,L-谷氨酸、L-苯丙氨酸、L-酪氨酸和L-色氨酸等多种氨基酸存在显著差异,这些氨基酸对于认知功能至关重要。随后的差异分析和偏最小二乘判别分析(sPLS-DA)显示,随着子代的生长发育,这些代谢差异在脑组织中逐渐减少。KEGG通路分析揭示了不同发育阶段中,丙氨酸、天冬氨酸和谷氨酸代谢、谷胱甘肽代谢、精氨酸生物合成、氨酰tRNA生物合成、组氨酸代谢以及牛磺酸和亚牛磺酸代谢等多条通路的差异调控。Mantel检验和Spearman分析表明,MSD子代代谢物的变化可能与肠道微生物 Ruminococcus_1、 Ruminococcaceae_UCG-005和 Eubacterium_coprostanoligenes_group相关。生化分析进一步支持L-谷氨酸代谢途径的发育性变化。综上所述,这些发现表明,MSD不仅影响母体的健康,还对子代产生持久的代谢影响,尤其影响与认知功能相关的代谢途径。这凸显了关注母体睡眠健康的重要性,以减轻对后代潜在的长期影响。.
Keywords: Cognitive development; Glutamate metabolism; Maternal sleep deprivation; Metabolomics; Offspring.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
