Human parainfluenza virus 3 field strains undergo extracellular fusion protein cleavage to activate entry
- PMID: 39382296
- PMCID: PMC11559058
- DOI: 10.1128/mbio.02327-24
Human parainfluenza virus 3 field strains undergo extracellular fusion protein cleavage to activate entry
Abstract
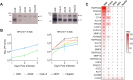
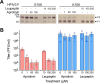
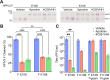
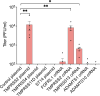
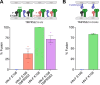
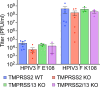
Human parainfluenza virus 3 (HPIV3) infection is driven by the coordinated action of viral surface glycoproteins hemagglutinin-neuraminidase (HN) and fusion protein (F). Receptor-engaged HN activates F to insert into the target cell membrane and drive virion-cell membrane fusion. For F to mediate entry, its precursor (F0) must first be cleaved by host proteases. F0 cleavage has been thought to be executed during viral glycoprotein transit through the trans-Golgi network by the ubiquitously expressed furin because F0 proteins of laboratory-adapted viruses contain a furin recognition dibasic cleavage motif RXKR around residue 108. Here, we show that the F proteins of field strains have a different cleavage motif from laboratory-adapted strains and are cleaved by unidentified proteases expressed in only a narrow subset of cell types. We demonstrate that extracellular serine protease inhibitors block HPIV3 F0 cleavage for field strains, suggesting F0 cleavage occurs at the cell surface facilitated by transmembrane proteases. Candidate proteases that may process HPIV3 F in vivo were identified by a genome-wide CRISPRa screen in HEK293/dCas9-VP64 + MPH cells. The lung-expressed extracellular serine proteases TMPRSS2 and TMPRSS13 are both sufficient to cleave HPIV3 F and enable infectious virus release by otherwise non-permissive cells. Our findings support an alternative mechanism of F activation in vivo, reliant on extracellular membrane-bound serine proteases expressed in a narrow subset of cells. The proportion of HPIV3 F proteins cleaved and infectious virus release is determined by host cell expression of requisite proteases, allowing just-in-time activation of F and positioning F cleavage as another key regulator of HPIV3 spread.
Importance: Enveloped viruses cause a wide range of diseases in humans. At the first step of infection, these viruses must fuse their envelope with a cell membrane to initiate infection. This fusion is mediated by viral proteins that require a critical activating cleavage event. It was previously thought that for parainfluenza virus 3, an important cause of respiratory disease and a representative of a group of important pathogens, this cleavage event was mediated by furin in the cell secretory pathways prior to formation of the virions. We show that this is only true for laboratory strain viruses, and that clinical viruses that infect humans utilize extracellular proteases that are only made by a small subset of cells. These results highlight the importance of studying authentic clinical viruses that infect human tissues for understanding natural infection.
Keywords: fusion protein; membrane fusion; parainfluenza virus; paramyxovirus; proteases; viral entry.
Conflict of interest statement
K.S., M.P., and A.M. are listed as inventors on a provisional patent related to the work in this article.
Figures


References
-
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simões EAF, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1 - DOI - PMC - PubMed
-
- Weinberg GA, Hall CB, Iwane MK, Poehling KA, Edwards KM, Griffin MR, Staat MA, Curns AT, Erdman DD, Szilagyi PG, New Vaccine Surveillance Network . 2009. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. J Pediatr 154:694–699. doi: 10.1016/j.jpeds.2008.11.034 - DOI - PubMed
-
- Black RE, Cousens S, Johnson HL, Lawn JE, Rudan I, Bassani DG, Jha P, Campbell H, Walker CF, Cibulskis R, Eisele T, Liu L, Mathers C. 2010. Global, regional, and national causes of child mortality in 2008: a systematic analysis. The Lancet 375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI114736/AI/NIAID NIH HHS/United States
- R01AI031971, R01AI114736, R01AI175362/HHS | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- F31 AI176760/AI/NIAID NIH HHS/United States
- R01 AI175362/AI/NIAID NIH HHS/United States
- Sharon Golub Fund at Columbia University Vagelos College of Physicians & Surgeons
LinkOut - more resources
Full Text Sources

