Robust estimation of cancer and immune cell-type proportions from bulk tumor ATAC-Seq data
- PMID: 39383060
- PMCID: PMC11464006
- DOI: 10.7554/eLife.94833
Robust estimation of cancer and immune cell-type proportions from bulk tumor ATAC-Seq data
Abstract
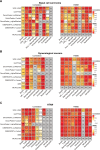
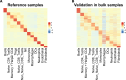
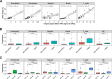
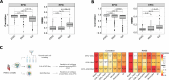
Assay for Transposase-Accessible Chromatin sequencing (ATAC-Seq) is a widely used technique to explore gene regulatory mechanisms. For most ATAC-Seq data from healthy and diseased tissues such as tumors, chromatin accessibility measurement represents a mixed signal from multiple cell types. In this work, we derive reliable chromatin accessibility marker peaks and reference profiles for most non-malignant cell types frequently observed in the microenvironment of human tumors. We then integrate these data into the EPIC deconvolution framework (Racle et al., 2017) to quantify cell-type heterogeneity in bulk ATAC-Seq data. Our EPIC-ATAC tool accurately predicts non-malignant and malignant cell fractions in tumor samples. When applied to a human breast cancer cohort, EPIC-ATAC accurately infers the immune contexture of the main breast cancer subtypes.
Keywords: ATAC-Seq; bulk deconvolution; cancer biology; chromatin accessibility; computational biology; human; systems biology; tumor microenvironment.
© 2024, Gabriel et al.
Conflict of interest statement
AG, JR, MF, CJ No competing interests declared, DG has recieved consulting fees from CeCaVa and Gnubiotics
Figures













Update of
- doi: 10.1101/2023.10.11.561826
- doi: 10.7554/eLife.94833.1
- doi: 10.7554/eLife.94833.2
- doi: 10.7554/eLife.94833.3
References
-
- 10x Genomics PBMC from a healthy donor - granulocytes removed through cell sorting (10k) 2021. [September 6, 2023]. https://www.10xgenomics.com/resources/datasets/pbmc-from-a-healthy-donor...
-
- Abascal F, Acosta R, Addleman NJ, Adrian J, Afzal V, Ai R, Aken B, Akiyama JA, Jammal OA, Amrhein H, Anderson SM, Andrews GR, Antoshechkin I, Ardlie KG, Armstrong J, Astley M, Banerjee B, Barkal AA, Barnes IHA, Barozzi I, Barrell D, Barson G, Bates D, Baymuradov UK, Bazile C, Beer MA, Beik S, Bender MA, Bennett R, Bouvrette LPB, Bernstein BE, Berry A, Bhaskar A, Bignell A, Blue SM, Bodine DM, Boix C, Boley N, Borrman T, Borsari B, Boyle AP, Brandsmeier LA, Breschi A, Bresnick EH, Brooks JA, Buckley M, Burge CB, Byron R, Cahill E, Cai L, Cao L, Carty M, Castanon RG, Castillo A, Chaib H, Chan ET, Chee DR, Chee S, Chen H, Chen H, Chen J-Y, Chen S, Cherry JM, Chhetri SB, Choudhary JS, Chrast J, Chung D, Clarke D, Cody NAL, Coppola CJ, Coursen J, D’Ippolito AM, Dalton S, Danyko C, Davidson C, Davila-Velderrain J, Davis CA, Dekker J, Deran A, DeSalvo G, Despacio-Reyes G, Dewey CN, Dickel DE, Diegel M, Diekhans M, Dileep V, Ding B, Djebali S, Dobin A, Dominguez D, Donaldson S, Drenkow J, Dreszer TR, Drier Y, Duff MO, Dunn D, Eastman C, Ecker JR, Edwards MD, El-Ali N, Elhajjajy SI, Elkins K, Emili A, Epstein CB, Evans RC, Ezkurdia I, Fan K, Farnham PJ, Farrell NP, Feingold EA, Ferreira A-M, Fisher-Aylor K, Fitzgerald S, Flicek P, Foo CS, Fortier K, Frankish A, Freese P, Fu S, Fu X-D, Fu Y, Fukuda-Yuzawa Y, Fulciniti M, Funnell APW, Gabdank I, Galeev T, Gao M, Giron CG, Garvin TH, Gelboin-Burkhart CA, Georgolopoulos G, Gerstein MB, Giardine BM, Gifford DK, Gilbert DM, Gilchrist DA, Gillespie S, Gingeras TR, Gong P, Gonzalez A, Gonzalez JM, Good P, Goren A, Gorkin DU, Graveley BR, Gray M, Greenblatt JF, Griffiths E, Groudine MT, Grubert F, Gu M, Guigó R, Guo H, Guo Y, Guo Y, Gursoy G, Gutierrez-Arcelus M, Halow J, Hardison RC, Hardy M, Hariharan M, Harmanci A, Harrington A, Harrow JL, Hashimoto TB, Hasz RD, Hatan M, Haugen E, Hayes JE, He P, He Y, Heidari N, Hendrickson D, Heuston EF, Hilton JA, Hitz BC, Hochman A, Holgren C, Hou L, Hou S, Hsiao Y-HE, Hsu S, Huang H, Hubbard TJ, Huey J, Hughes TR, Hunt T, Ibarrientos S, Issner R, Iwata M, Izuogu O, Jaakkola T, Jameel N, Jansen C, Jiang L, Jiang P, Johnson A, Johnson R, Jungreis I, Kadaba M, Kasowski M, Kasparian M, Kato M, Kaul R, Kawli T, Kay M, Keen JC, Keles S, Keller CA, Kelley D, Kellis M, Kheradpour P, Kim DS, Kirilusha A, Klein RJ, Knoechel B, Kuan S, Kulik MJ, Kumar S, Kundaje A, Kutyavin T, Lagarde J, Lajoie BR, Lambert NJ, Lazar J, Lee AY, Lee D, Lee E, Lee JW, Lee K, Leslie CS, Levy S, Li B, Li H, Li N, Li S, Li X, Li YI, Li Y, Li Y, Li Y, Lian J, Libbrecht MW, Lin S, Lin Y, Liu D, Liu J, Liu P, Liu T, Liu XS, Liu Y, Liu Y, Long M, Lou S, Loveland J, Lu A, Lu Y, Lécuyer E, Ma L, Mackiewicz M, Mannion BJ, Mannstadt M, Manthravadi D, Marinov GK, Martin FJ, Mattei E, McCue K, McEown M, McVicker G, Meadows SK, Meissner A, Mendenhall EM, Messer CL, Meuleman W, Meyer C, Miller S, Milton MG, Mishra T, Moore DE, Moore HM, Moore JE, Moore SH, Moran J, Mortazavi A, Mudge JM, Munshi N, Murad R, Myers RM, Nandakumar V, Nandi P, Narasimha AM, Narayanan AK, Naughton H, Navarro FCP, Navas P, Nazarovs J, Nelson J, Neph S, Neri FJ, Nery JR, Nesmith AR, Newberry JS, Newberry KM, Ngo V, Nguyen R, Nguyen TB, Nguyen T, Nishida A, Noble WS, Novak CS, Novoa EM, Nuñez B, O’Donnell CW, Olson S, Onate KC, Otterman E, Ozadam H, Pagan M, Palden T, Pan X, Park Y, Partridge EC, Paten B, Pauli-Behn F, Pazin MJ, Pei B, Pennacchio LA, Perez AR, Perry EH, Pervouchine DD, Phalke NN, Pham Q, Phanstiel DH, Plajzer-Frick I, Pratt GA, Pratt HE, Preissl S, Pritchard JK, Pritykin Y, Purcaro MJ, Qin Q, Quinones-Valdez G, Rabano I, Radovani E, Raj A, Rajagopal N, Ram O, Ramirez L, Ramirez RN, Rausch D, Raychaudhuri S, Raymond J, Razavi R, Reddy TE, Reimonn TM, Ren B, Reymond A, Reynolds A, Rhie SK, Rinn J, Rivera M, Rivera-Mulia JC, Roberts BS, Rodriguez JM, Rozowsky J, Ryan R, Rynes E, Salins DN, Sandstrom R, Sasaki T, Sathe S, Savic D, Scavelli A, Scheiman J, Schlaffner C, Schloss JA, Schmitges FW, See LH, Sethi A, Setty M, Shafer A, Shan S, Sharon E, Shen Q, Shen Y, Sherwood RI, Shi M, Shin S, Shoresh N, Siebenthall K, Sisu C, Slifer T, Sloan CA, Smith A, Snetkova V, Snyder MP, Spacek DV, Srinivasan S, Srivas R, Stamatoyannopoulos G, Stamatoyannopoulos JA, Stanton R, Steffan D, Stehling-Sun S, Strattan JS, Su A, Sundararaman B, Suner M-M, Syed T, Szynkarek M, Tanaka FY, Tenen D, Teng M, Thomas JA, Toffey D, Tress ML, Trout DE, Trynka G, Tsuji J, Upchurch SA, Ursu O, Uszczynska-Ratajczak B, Uziel MC, Valencia A, Biber BV, van der Velde AG, Van Nostrand EL, Vaydylevich Y, Vazquez J, Victorsen A, Vielmetter J, Vierstra J, Visel A, Vlasova A, Vockley CM, Volpi S, Vong S, Wang H, Wang M, Wang Q, Wang R, Wang T, Wang W, Wang X, Wang Y, Watson NK, Wei X, Wei Z, Weisser H, Weissman SM, Welch R, Welikson RE, Weng Z, Westra H-J, Whitaker JW, White C, White KP, Wildberg A, Williams BA, Wine D, Witt HN, Wold B, Wolf M, Wright J, Xiao R, Xiao X, Xu J, Xu J, Yan K-K, Yan Y, Yang H, Yang X, Yang Y-W, Yardımcı GG, Yee BA, Yeo GW, Young T, Yu T, Yue F, Zaleski C, Zang C, Zeng H, Zeng W, Zerbino DR, Zhai J, Zhan L, Zhan Y, Zhang B, Zhang J, Zhang J, Zhang K, Zhang L, Zhang P, Zhang Q, Zhang X-O, Zhang Y, Zhang Z, Zhao Y, Zheng Y, Zhong G, Zhou X-Q, Zhu Y, Zimmerman J, Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, Kawli T, Davis CA, Dobin A, Kaul R, Halow J, Van Nostrand EL, Freese P, Gorkin DU, Shen Y, He Y, Mackiewicz M, Pauli-Behn F, Williams BA, Mortazavi A, Keller CA, Zhang X-O, Elhajjajy SI, Huey J, Dickel DE, Snetkova V, Wei X, Wang X, Rivera-Mulia JC, Rozowsky J, Zhang J, Chhetri SB, Zhang J, Victorsen A, White KP, Visel A, Yeo GW, Burge CB, Lécuyer E, Gilbert DM, Dekker J, Rinn J, Mendenhall EM, Ecker JR, Kellis M, Klein RJ, Noble WS, Kundaje A, Guigó R, Farnham PJ, Cherry JM, Myers RM, Ren B, Graveley BR, Gerstein MB, Pennacchio LA, Snyder MP, Bernstein BE, Wold B, Hardison RC, Gingeras TR, Stamatoyannopoulos JA, Weng Z, The ENCODE Project Consortium Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583:699–710. doi: 10.1038/s41586-020-2493-4. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.21992609.v1
LinkOut - more resources
Full Text Sources
Medical

