Epigenetic regulation of p63 blocks squamous-to-neuroendocrine transdifferentiation in esophageal development and malignancy
- PMID: 39383220
- PMCID: PMC11463268
- DOI: 10.1126/sciadv.adq0479
Epigenetic regulation of p63 blocks squamous-to-neuroendocrine transdifferentiation in esophageal development and malignancy
Abstract
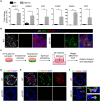
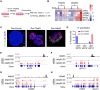
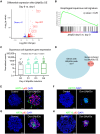
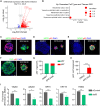
While cell fate determination and maintenance are important in establishing and preserving tissue identity and function during development, aberrant cell fate transition leads to cancer cell heterogeneity and resistance to treatment. Here, we report an unexpected role for the transcription factor p63 (Trp63/TP63) in the fate choice of the squamous versus neuroendocrine lineage in esophageal development and malignancy. Deletion of p63 results in extensive neuroendocrine differentiation in the developing mouse esophagus and esophageal progenitors derived from human embryonic stem cells. In human esophageal neuroendocrine carcinoma (eNEC) cells, p63 is transcriptionally silenced by EZH2-mediated H3K27 trimethylation (H3K27me3). Up-regulation of the major p63 isoform ΔNp63α, through either ectopic expression or EZH2 inhibition, promotes squamous transdifferentiation of eNEC cells. Together, these findings uncover p63 as a rheostat in coordinating the transition between squamous and neuroendocrine cell fates during esophageal development and tumor progression.
Figures



Update of
-
Epigenetic regulation of p63 blocks squamous-to-neuroendocrine transdifferentiation in esophageal development and malignancy.bioRxiv [Preprint]. 2023 Sep 11:2023.09.09.556982. doi: 10.1101/2023.09.09.556982. bioRxiv. 2023. Update in: Sci Adv. 2024 Oct 11;10(41):eadq0479. doi: 10.1126/sciadv.adq0479. PMID: 37745439 Free PMC article. Updated. Preprint.
References
-
- Jabbari M., Goresky C. A., Lough J., Yaffe C., Daly D., Cote C., The inlet patch: Heterotopic gastric mucosa in the upper esophagus. Gastroenterology 89, 352–356 (1985). - PubMed
-
- Spechler S. J., Zeroogian J. M., Antonioli D. A., Wang H. H., Goyal R. K., Prevalence of metaplasia at the gastro-oesophageal junction. Lancet 344, 1533–1536 (1994). - PubMed
-
- Jiang M., Li H., Zhang Y., Yang Y., Lu R., Liu K., Lin S., Lan X., Wang H., Wu H., Zhu J., Zhou Z., Xu J., Lee D. K., Zhang L., Lee Y. C., Yuan J., Abrams J. A., Wang T. C., Sepulveda A. R., Wu Q., Chen H., Sun X., She J., Chen X., Que J., Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature 550, 529–533 (2017). - PMC - PubMed
-
- Quante M., Bhagat G., Abrams J. A., Marache F., Good P., Lee M. D., Lee Y., Friedman R., Asfaha S., Dubeykovskaya Z., Mahmood U., Figueiredo J. L., Kitajewski J., Shawber C., Lightdale C. J., Rustgi A. K., Wang T. C., Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell 21, 36–51 (2012). - PMC - PubMed
-
- Braslis K. G., David R. C., Nelson E., Civantos F., Soloway M. S., Squamous cell carcinoma of the prostate: A transformation from adenocarcinoma after the use of a luteinizing hormone-releasing hormone agonist and flutamide. Urology 45, 329–331 (1995). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 DK132251/DK/NIDDK NIH HHS/United States
- R01 CA272901/CA/NCI NIH HHS/United States
- P30 DK132710/DK/NIDDK NIH HHS/United States
- S10 OD020056/OD/NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R01 DK120650/DK/NIDDK NIH HHS/United States
- IK6 BX006300/BX/BLRD VA/United States
- I01 BX002115/BX/BLRD VA/United States
- P01 CA268991/CA/NCI NIH HHS/United States
- R01 AR073170/AR/NIAMS NIH HHS/United States
- R01 DE031873/DE/NIDCR NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
- S10 OD032447/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

