VGLL3 modulates chemosensitivity through promoting DNA double-strand break repair
- PMID: 39383226
- PMCID: PMC11463272
- DOI: 10.1126/sciadv.adr2643
VGLL3 modulates chemosensitivity through promoting DNA double-strand break repair
Abstract
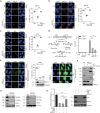
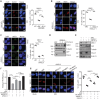
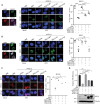
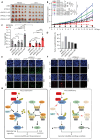
Transcription cofactor vestigial-like 3 (VGLL3), as a master regulator of female-biased autoimmunity, also functions in tumor development, while the underlying mechanisms remain largely elusive. Here, we report that VGLL3 plays an important role in DNA damage response (DDR). VGLL3 can be recruited to damage sites in a PARylation-dependent manner. VGLL3 depletion impairs the accumulation of RNF8 and RAD51 at sites of DNA damage, leading to reduced homologous recombination efficiency and increased cellular sensitivity to chemotherapeutic drugs. Mechanistically, VGLL3 can prevent CtIP from KLHL15-mediated ubiquitination and degradation through competitive binding with KLHL15 and, meanwhile, stabilize MDC1 by limiting TRIP12-MDC1 but promoting USP7-MDC1 associations for optimal RNF8 signaling initiation. Consistently, VGLL3 depletion delays tumor development and sensitizes the xenografts to etoposide treatment. Overall, our results reveal an unexpected role of VGLL3 in DDR, which is distinct from its transcriptional cofactor function and not conserved among VGLL family members.
Figures




References
-
- Horii Y., Matsuda S., Toyota C., Morinaga T., Nakaya T., Tsuchiya S., Ohmuraya M., Hironaka T., Yoshiki R., Kasai K., Yamauchi Y., Takizawa N., Nagasaka A., Tanaka A., Kosako H., Nakaya M., VGLL3 is a mechanosensitive protein that promotes cardiac fibrosis through liquid-liquid phase separation. Nat. Commun. 14, 550 (2023). - PMC - PubMed
-
- Liang Y., Tsoi L. C., Xing X., Beamer M. A., Swindell W. R., Sarkar M. K., Berthier C. C., Stuart P. E., Harms P. W., Nair R. P., Elder J. T., Voorhees J. J., Kahlenberg J. M., Gudjonsson J. E., A gene network regulated by the transcription factor VGLL3 as a promoter of sex-biased autoimmune diseases. Nat. Immunol. 18, 152–160 (2017). - PMC - PubMed
-
- Billi A. C., Gharaee-Kermani M., Fullmer J., Tsoi L. C., Hill B. D., Gruszka D., Ludwig J., Xing X., Estadt S., Wolf S. J., Rizvi S. M., Berthier C. C., Hodgin J. B., Beamer M. A., Sarkar M. K., Liang Y., Uppala R., Shao S., Zeng C., Harms P. W., Verhaegen M. E., Voorhees J. J., Wen F., Ward N. L., Dlugosz A. A., Kahlenberg J. M., Gudjonsson J. E., The female-biased factor VGLL3 drives cutaneous and systemic autoimmunity. JCI Insight 4, e127291 (2019). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

