A proinflammatory stem cell niche drives myelofibrosis through a targetable galectin-1 axis
- PMID: 39383242
- PMCID: PMC7616771
- DOI: 10.1126/scitranslmed.adj7552
A proinflammatory stem cell niche drives myelofibrosis through a targetable galectin-1 axis
Abstract
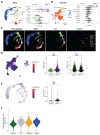
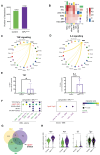
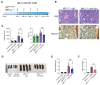
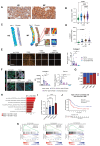
Myeloproliferative neoplasms are stem cell-driven cancers associated with a large burden of morbidity and mortality. Most patients present with early-stage disease, but a substantial proportion progress to myelofibrosis or secondary leukemia, advanced cancers with a poor prognosis and high symptom burden. Currently, it remains difficult to predict progression, and therapies that reliably prevent or reverse fibrosis are lacking. A major bottleneck to the discovery of disease-modifying therapies has been an incomplete understanding of the interplay between perturbed cellular and molecular states. Several cell types have individually been implicated, but a comprehensive analysis of myelofibrotic bone marrow is lacking. We therefore mapped the cross-talk between bone marrow cell types in myelofibrotic bone marrow. We found that inflammation and fibrosis are orchestrated by a "quartet" of immune and stromal cell lineages, with basophils and mast cells creating a TNF signaling hub, communicating with megakaryocytes, mesenchymal stromal cells, and proinflammatory fibroblasts. We identified the β-galactoside-binding protein galectin-1 as a biomarker of progression to myelofibrosis and poor survival in multiple patient cohorts and as a promising therapeutic target, with reduced myeloproliferation and fibrosis in vitro and in vivo and improved survival after galectin-1 inhibition. In human bone marrow organoids, TNF increased galectin-1 expression, suggesting a feedback loop wherein the proinflammatory myeloproliferative neoplasm clone creates a self-reinforcing niche, fueling progression to advanced disease. This study provides a resource for studying hematopoietic cell-niche interactions, with relevance for cancer-associated inflammation and disorders of tissue fibrosis.
Conflict of interest statement
B. Psaila: Alethiomics (co-founder, equity holder, consultancy, research funding), Incyte (research funding, consultancy), Constellation Therapeutics (consultancy), Blueprint Medicines (advisory board, consultancy), Galecto (research funding), Novartis (paid speaking engagements, consultancy); GSK (consultancy, paid speaking engagements), BMS (consultancy). AJM has received honoraria for consulting and speaker fees from Novartis, Celgene/BMS, Abbvie, CTI, MD-Education, Sierra Oncology, Medialis, Morphosys, Ionis, Mescape, Karyopharm, Sensyn, Incyte, Galecto, Pfizer, Relay Therapeutics, GSK, Alethiomics & Gilead. AJM has received research funding from Celgene/BMS, Novartis, Roche, Alethiomics and Galecto. AJM is co-founder and equity holder in Alethiomics Ltd, a spin out company from the University of Oxford. A.O. Khan: Alethiomics (consultancy). Two patents have been filed by A.O. Khan and B. Psaila relating to the human bone marrow organoids platform utilised in this manuscript (GB2202025.9, GB2216647 and 2402478.8).
Figures



References
-
- Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, Chen W, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia. 2022;36:1703–1719. doi: 10.1038/s41375-022-01613-1. - DOI - PMC - PubMed
-
- Koschmieder S, Mughal TI, Hasselbalch HC, Barosi G, Valent P, Kiladjian JJ, Jeryczynski G, Gisslinger H, Jutzi JS, Pahl HL, Hehlmann R, et al. Myeloproliferative neoplasms and inflammation: whether to target the malignant clone or the inflammatory process or both. Leukemia. 2016;30:1018–1024. - PubMed
-
- Koschmieder S, Chatain N. Role of inflammation in the biology of myeloproliferative neoplasms. Blood Rev. 2020;42:100711. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

