Binding kinetics of quaternary ammonium ions in Kcv potassium channels
- PMID: 39383513
- PMCID: PMC11575739
- DOI: 10.1080/19336950.2024.2402749
Binding kinetics of quaternary ammonium ions in Kcv potassium channels
Abstract
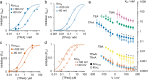
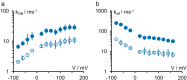
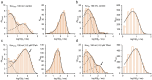
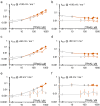
Kcv channels from plant viruses represent the autonomous pore module of potassium channels, devoid of any regulatory domains. These small proteins show very reproducible single-channel behavior in planar lipid bilayers. Thus, they are an optimum system for the study of the biophysics of ion transport and gating. Structural models based on homology modeling have been used successfully, but experimental structural data are currently not available. Here we determine the size of the cytosolic pore entrance by studying the blocker kinetics. Blocker binding and dissociation rate constants ranging from 0.01 to 1000 ms-1 were determined for different quaternary ammonium ions. We found that the cytosolic pore entrance of KcvNTS must be at least 11 Å wide. The results further indicate that the residues controlling a cytosolic gate in one of the Kcv isoforms influence blocker binding/dissociation as well as a second gate even when the cytosolic gate is in the open state. The voltage dependence of the rate constant of blocker release is used to test, which blockers bind to the same binding site.
Keywords: Fast block; blocker; diffusion limitation; planar lipid bilayer; single-channel currents; viral potassium channel.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
