Up-regulated Lnc BTU promotes the production of duck plague virus DNA polymerase and inhibits the activation of JAK-STAT pathway to facilitate duck plague virus replication
- PMID: 39383668
- PMCID: PMC11490923
- DOI: 10.1016/j.psj.2024.104238
Up-regulated Lnc BTU promotes the production of duck plague virus DNA polymerase and inhibits the activation of JAK-STAT pathway to facilitate duck plague virus replication
Abstract
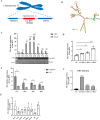
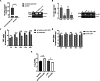
Duck plague virus (DPV) is the only herpes virus known to be transmissible among aquatic animals, leading to immunosuppression in ducks, geese and swans. Long noncoding RNAs (LncRNA) are known to participate in viral infections, acting as either immune defenders or viral targets to evade the host response, but their precise roles in waterfowl virus infections are yet to be fully understood. This study aimed to investigate the role of LncRNA in DPV-induced innate immune responses. Results showed that DPV infection greatly upregulated Lnc BTU expression in duck embryo fibroblasts (DEF) and Lnc BTU promoted DPV replication. Mechanically, 4 DPV proteins, namely UL46, UL42, VP22 and US10, interacted with Lnc BTU, leading to its upregulation. Specifically, Lnc BTU facilitated the production of DNA polymerase by enhancing UL42 expression, thereby promoting DPV replication. Additionally, Lnc BTU suppressed STAT1 expression by targeting the DNA binding domain (DBD) and promoting STAT1 degradation through the proteasome pathway. Furthermore, Lnc BTU inhibited the production of key antiviral factors such as IFN-α, IFN-β, MX and OASL during DPV infection. Treatment with 2 JAK-STAT pathway activators in DEFs resulted in the inhibition of Lnc BTU expression and DPV replication. Interestingly, DPV infection led to a decrease in STAT1 levels, which was reversed by Si-Lnc BTU. These findings suggest that DPV relies on Lnc BTU to inhibit the activation of the JAK-STAT pathway and limit the production of type 1 interferons (IFN) to complete immune evasion. Our study highlights the novel role of DPV proteins UL46, UL42, VP22, US10 as RNA-binding proteins in modulating the innate antiviral immune response, and discover the role of a new host factor, Lnc BTU, in DPV immune evasion, Lnc BTU and STAT1 can be used as a potential therapeutic target for DPV infection and immune evasion.
Keywords: Duck plague virus; JAK-STAT pathway; Lnc BTU; STAT1; innate immunity.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DISCLOSURES The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Apinda N., Muenthaisong A., Chomjit P., Sangkakam K., Nambooppha B., Rittipornlertrak A., Koonyosying P., Yao Y., Nair V., Sthitmatee N. Simultaneous protective immune responses of Ducks against Duck plague and fowl cholera by recombinant duck enteritis virus vector expressing pasteurella multocida OmpH gene. Vaccines. 2022;10:1358. - PMC - PubMed
-
- Burrel S., Aït-Arkoub Z., Agut H., Boutolleau D. Genotypic characterization of herpes simplex virus DNA polymerase UL42 processivity factor. Antiviral. Res. 2012;93:199–203. - PubMed
-
- Cai B., Cai J., Luo Y., Chen C., Zhang S. The Specific roles of JAK/STAT signaling pathway in sepsis. Inflammation. 2015;38:1599–1608. - PubMed
-
- Cai R., Sun Y., Qimuge N., Wang G., Wang Y., Chu G., Yu T., Yang G., Pang W. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating adiponectin mRNA translation. Biochim. Biophys. Acta. Mol. Cell. Biol. Lipids. 2018;1863:420–432. - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

