A human lectin array for characterizing host-pathogen interactions
- PMID: 39384043
- PMCID: PMC11566865
- DOI: 10.1016/j.jbc.2024.107869
A human lectin array for characterizing host-pathogen interactions
Abstract
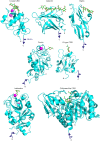
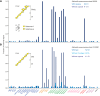
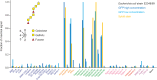
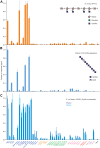
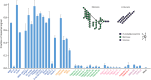
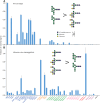
A human lectin array has been developed to probe the interactions of innate immune receptors with pathogenic and commensal microorganisms. Following the successful introduction of a lectin array containing all of the cow C-type carbohydrate-recognition domains (CRDs), a human array described here contains the C-type CRDs as well as CRDs from other classes of sugar-binding receptors, including galectins, siglecs, R-type CRDs, ficolins, intelectins, and chitinase-like lectins. The array is constructed with CRDs modified with single-site biotin tags, ensuring that the sugar-binding sites in CRDs are displayed on a streptavidin-coated surface in a defined orientation and are accessible to the surfaces of microbes. A common approach used for expression and display of CRDs from all of the different structural categories of glycan-binding receptors allows comparisons across lectin families. In addition to previously documented protocols for binding of fluorescently labeled bacteria, methods have been developed for detecting unlabeled bacteria bound to the array by counter-staining with DNA-binding dye. Screening has also been undertaken with viral glycoproteins and bacterial and fungal polysaccharides. The array provides an unbiased screen for sugar ligands that interact with receptors and many show binding not anticipated from earlier studies. For example, some of the galectins bind with high affinity to bacterial glycans that lack lactose or N-acetyllactosamine. The results demonstrate the utility of the human lectin array for providing a unique overview of the interactions of multiple classes of glycan-binding proteins in the innate immune system with different types of microorganisms.
Keywords: array screening; carbohydrate-binding protein; glycan-binding receptors; glycobiology; host-pathogen interaction; innate immunity; lectin.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures
References
-
- Li Z., Feizi T. The neoglycolipid (NGL) technology-based microarrays and future prospects. FEBS Lett. 2018;592:3976–3991. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

