Molecular characterization and biomarker identification in paediatric B-cell acute lymphoblastic leukaemia
- PMID: 39384181
- PMCID: PMC11464031
- DOI: 10.1111/jcmm.70126
Molecular characterization and biomarker identification in paediatric B-cell acute lymphoblastic leukaemia
Abstract
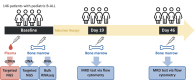
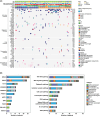
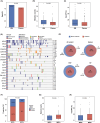
B-cell acute lymphoblastic leukaemia (B-ALL) is the most prevalent hematologic malignancy in children and a leading cause of mortality. Managing B-ALL remains challenging due to its heterogeneity and relapse risk. This study aimed to delineate the molecular features of paediatric B-ALL and explore the clinical utility of circulating tumour DNA (ctDNA). We analysed 146 patients with paediatric B-ALL who received systemic chemotherapy. The mutational landscape was profiled in bone marrow (BM) and plasma samples using next-generation sequencing. Minimal residual disease (MRD) testing on day 19 of induction therapy evaluated treatment efficacy. RNA sequencing identified gene fusions in 61% of patients, including 37 novel fusions. Specifically, the KMT2A-TRIM29 novel fusion was validated in a boy who responded well to initial therapy but relapsed after 1 year. Elevated mutation counts and maximum variant allele frequency in baseline BM were associated with significantly poorer chemotherapy response (p = 0.0012 and 0.028, respectively). MRD-negative patients exhibited upregulation of immune-related pathways (p < 0.01) and increased CD8+ T cell infiltration (p = 0.047). Baseline plasma ctDNA exhibited high mutational concordance with the paired BM samples and was significantly associated with chemotherapy efficacy. These findings suggest that ctDNA and BM profiling offer promising prognostic insights for paediatric B-ALL management.
Keywords: ctDNA; immune microenvironment; next‐generation sequencing; paediatric B‐cell acute lymphoblastic leukaemia; predictive biomarker.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors confirm that there are no conflicts of interest.
Figures


References
-
- Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541‐1552. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

