De novo production of protoberberine and benzophenanthridine alkaloids through metabolic engineering of yeast
- PMID: 39384562
- PMCID: PMC11464499
- DOI: 10.1038/s41467-024-53045-3
De novo production of protoberberine and benzophenanthridine alkaloids through metabolic engineering of yeast
Abstract
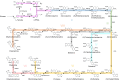
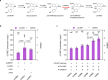
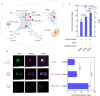
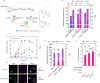
Protoberberine alkaloids and benzophenanthridine alkaloids (BZDAs) are subgroups of benzylisoquinoline alkaloids (BIAs), which represent a diverse class of plant-specialized natural metabolites with many pharmacological properties. Microbial biosynthesis has been allowed for accessibility and scalable production of high-value BIAs. Here, we engineer Saccharomyces cerevisiae to de novo produce a series of protoberberines and BZDAs, including palmatine, berberine, chelerythrine, sanguinarine and chelirubine. An ER compartmentalization strategy is developed to improve vacuole protein berberine bridge enzyme (BBE) activity, resulting in >200% increase on the production of the key intermediate (S)-scoulerine. Another promiscuous vacuole protein dihydrobenzophenanthridine oxidase (DBOX) has been identified to catalyze two-electron oxidation on various tetrahydroprotoberberines at N7-C8 position and dihydrobenzophenanthridine alkaloids. Furthermore, cytosolically expressed DBOX can alleviate the limitation on BBE. This study highlights the potential of microbial cell factories for the biosynthesis of a diverse group of BIAs through engineering of heterologous plant enzymes.
© 2024. The Author(s).
Conflict of interest statement
Y.C. and X.J. are inventors of pending patent applications (PCT/071270 and PCT/071276) arising from work on strategies for improved alkaloids production. Other authors declare no competing interests.
Figures


References
-
- World Health Organization. Global action plan on antimicrobial resistance. (Jan 2016). - PubMed
-
- Schmeller, T., Latz-Brüning, B. & Wink, M. Biochemical activities of berberine, palmatine and sanguinarine mediating chemical defence against microorganisms and herbivores. Phytochemistry44, 257–266 (1997). - PubMed
-
- Liscombe, D. K. & Facchini, P. J. Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Curr. Opin. Biotechnol.19, 173–180 (2008). - PubMed
-
- Imenshahidi, M. & Hosseinzadeh, H. Berberis vulgaris and berberine: an update review. Phytother. Res.30, 1745–1764 (2016). - PubMed
-
- Derosa, G., Maffioli, P. & Arrigo, F. G. C. Berberine on metabolic and cardiovascular risk factors: an analysis from preclinical evidences to clinical trials. Expert Opin. Biol. Ther.12, 8 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

