Elucidation of Spartina dimethylsulfoniopropionate synthesis genes enables engineering of stress tolerant plants
- PMID: 39384757
- PMCID: PMC11464771
- DOI: 10.1038/s41467-024-51758-z
Elucidation of Spartina dimethylsulfoniopropionate synthesis genes enables engineering of stress tolerant plants
Abstract
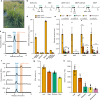
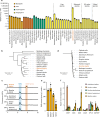
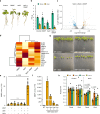
The organosulfur compound dimethylsulfoniopropionate (DMSP) has key roles in stress protection, global carbon and sulfur cycling, chemotaxis, and is a major source of climate-active gases. Saltmarshes are global hotspots for DMSP cycling due to Spartina cordgrasses that produce exceptionally high concentrations of DMSP. Here, in Spartina anglica, we identify the plant genes that underpin high-level DMSP synthesis: methionine S-methyltransferase (MMT), S-methylmethionine decarboxylase (SDC) and DMSP-amine oxidase (DOX). Homologs of these enzymes are common in plants, but differences in expression and catalytic efficiency explain why S. anglica accumulates such high DMSP concentrations and other plants only accumulate low concentrations. Furthermore, DMSP accumulation in S. anglica is consistent with DMSP having a role in oxidative and osmotic stress protection. Importantly, administration of DMSP by root uptake or over-expression of Spartina DMSP synthesis genes confers plant tolerance to salinity and drought offering a route for future bioengineering for sustainable crop production.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Kirst, G. O. Salinity Tolerance of Eukaryotic Marine Algae. Annu. Rev. Plant Physiol. Plant Mol. Biol.41, 21–53 (1990). - DOI
-
- Karsten, U., Kück, K., Vogt, C. & Kirst, G. O. Dimethylsulfoniopropionate Production in Phototrophic Organisms and its Physiological Functions as a Cryoprotectant. in Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds (eds. Kiene, R. P., Visscher, P. T., Keller, M. D. & Kirst, G. O.) 143–153. 10.1007/978-1-4613-0377-0_13 (Springer US, Boston, MA, 1996).
-
- Wolfe, G. V. & Steinke, M. Grazing-activated production of dimethyl sulfide (DMS) by two clones of Emiliania huxleyi. Limnol. Oceanogr.41, 1151–1160 (1996). - DOI
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- NE/V000756/1/RCUK | Natural Environment Research Council (NERC)
- NE/V000756/1/RCUK | Natural Environment Research Council (NERC)
- NE/V000756/1/RCUK | Natural Environment Research Council (NERC)
- NE/S007334/1/RCUK | Natural Environment Research Council (NERC)
- MR/T044020/1/RCUK | Natural Environment Research Council (NERC)
- NE/V000756/1/RCUK | Natural Environment Research Council (NERC)
- NE/V000756/1/RCUK | Natural Environment Research Council (NERC)
- NE/P012671/RCUK | Natural Environment Research Council (NERC)
- NE/S001352/RCUK | Natural Environment Research Council (NERC)
- NE/X000990/RCUK | Natural Environment Research Council (NERC)
- NE/X014428/RCUK | Natural Environment Research Council (NERC)
- BB/X005968/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/X005968/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/X005968/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/T008717/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/T008717/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- BB/X005968/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- RPG-2020-413/Leverhulme Trust
LinkOut - more resources
Full Text Sources

