Multi-omics profiling of DNA methylation and gene expression alterations in human cocaine use disorder
- PMID: 39384764
- PMCID: PMC11464785
- DOI: 10.1038/s41398-024-03139-9
Multi-omics profiling of DNA methylation and gene expression alterations in human cocaine use disorder
Abstract
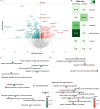
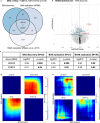
Structural and functional changes of the brain are assumed to contribute to excessive cocaine intake, craving, and relapse in cocaine use disorder (CUD). Epigenetic and transcriptional changes were hypothesized as a molecular basis for CUD-associated brain alterations. Here we performed a multi-omics study of CUD by integrating epigenome-wide methylomic (N = 42) and transcriptomic (N = 25) data from the same individuals using postmortem brain tissue of Brodmann Area 9 (BA9). Of the N = 1 057 differentially expressed genes (p < 0.05), one gene, ZFAND2A, was significantly upregulated in CUD at transcriptome-wide significance (q < 0.05). Differential alternative splicing (AS) analysis revealed N = 98 alternatively spliced transcripts enriched in axon and dendrite extension pathways. Strong convergent overlap in CUD-associated expression deregulation was found between our BA9 cohort and independent replication datasets. Epigenomic, transcriptomic, and AS changes in BA9 converged at two genes, ZBTB4 and INPP5E. In pathway analyses, synaptic signaling, neuron morphogenesis, and fatty acid metabolism emerged as the most prominently deregulated biological processes. Drug repositioning analysis revealed glucocorticoid receptor targeting drugs as most potent in reversing the CUD expression profile. Our study highlights the value of multi-omics approaches for an in-depth molecular characterization and provides insights into the relationship between CUD-associated epigenomic and transcriptomic signatures in the human prefrontal cortex.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Degenhardt L, Charlson F, Ferrari A, Santomauro D, Erskine H, Mantilla-Herrara A, et al. The global burden of disease attributable to alcohol and drug use in 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Psychiatry. 2018;5:987–1012. - DOI - PMC - PubMed
-
- American Psychiatric Association & American Psychiatric Association. DSM-5 Task Force. Diagnostic and statistical manual of mental disorders: DSM-5. Arlington: VAPA; 2013. .
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

