Structural basis for antibiotic transport and inhibition in PepT2
- PMID: 39384780
- PMCID: PMC11464717
- DOI: 10.1038/s41467-024-53096-6
Structural basis for antibiotic transport and inhibition in PepT2
Abstract
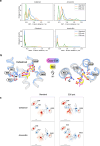
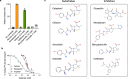
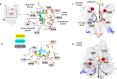
The uptake and elimination of beta-lactam antibiotics in the human body are facilitated by the proton-coupled peptide transporters PepT1 (SLC15A1) and PepT2 (SLC15A2). The mechanism by which SLC15 family transporters recognize and discriminate between different drug classes and dietary peptides remains unclear, hampering efforts to improve antibiotic pharmacokinetics through targeted drug design and delivery. Here, we present cryo-EM structures of the proton-coupled peptide transporter, PepT2 from Rattus norvegicus, in complex with the widely used beta-lactam antibiotics cefadroxil, amoxicillin and cloxacillin. Our structures, combined with pharmacophore mapping, molecular dynamics simulations and biochemical assays, establish the mechanism of beta-lactam antibiotic recognition and the important role of protonation in drug binding and transport.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Structural basis for antibiotic transport and inhibition in PepT2, the mammalian proton-coupled peptide transporter.Res Sq [Preprint]. 2024 May 30:rs.3.rs-4435259. doi: 10.21203/rs.3.rs-4435259/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Oct 9;15(1):8755. doi: 10.1038/s41467-024-53096-6. PMID: 38903084 Free PMC article. Updated. Preprint.
References
-
- Lax, E. The mold in Dr. Florey’s coat The story of the penicillin miracle, (Henry Holt and Co, New York, New York, USA, 2004).
-
- Livermore, D. M. & Woodford, N. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol14, 413–420 (2006). - PubMed
-
- Vollmer, W., Blanot, D. & de Pedro, M. A. Peptidoglycan structure and architecture. FEMS Microbiol Rev.32, 149–167 (2008). - PubMed
-
- Kato, K. et al. Intestinal absorption mechanism of tebipenem pivoxil, a novel oral carbapenem: involvement of human OATP family in apical membrane transport. Mol. pharmaceutics7, 1747–1756 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

