CBFA2T3-GLIS2 mediates transcriptional regulation of developmental pathways through a gene regulatory network
- PMID: 39384814
- PMCID: PMC11464917
- DOI: 10.1038/s41467-024-53158-9
CBFA2T3-GLIS2 mediates transcriptional regulation of developmental pathways through a gene regulatory network
Abstract
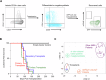
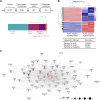
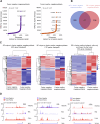
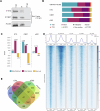
CBFA2T3-GLIS2 is a fusion oncogene found in pediatric acute megakaryoblastic leukemia that is associated with a poor prognosis. We establish a model of CBFA2T3-GLIS2 driven acute megakaryoblastic leukemia that allows the distinction of fusion specific changes from those that reflect the megakaryoblast lineage of this leukemia. Using this model, we map fusion genome wide binding that in turn imparts the characteristic transcriptional signature. A network of transcription factor genes bound and upregulated by the fusion are found to have downstream effects that result in dysregulated signaling of developmental pathways including NOTCH, Hedgehog, TGFβ, and WNT. Transcriptional regulation is mediated by homo-dimerization and binding of the ETO transcription factor through the nervy homology region 2. Loss of nerve homology region 2 abrogated the development of leukemia, leading to downregulation of JAK/STAT, Hedgehog, and NOTCH transcriptional signatures. These data contribute to the understanding of CBFA2T3-GLIS2 mediated leukemogenesis and identify potential therapeutic vulnerabilities for future studies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

