Single cell analysis revealed SFRP2 cancer associated fibroblasts drive tumorigenesis in head and neck squamous cell carcinoma
- PMID: 39384902
- PMCID: PMC11464629
- DOI: 10.1038/s41698-024-00716-5
Single cell analysis revealed SFRP2 cancer associated fibroblasts drive tumorigenesis in head and neck squamous cell carcinoma
Abstract
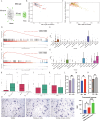
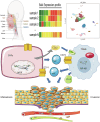
Understanding the mechanisms of invasion and metastasis in head and neck squamous cell carcinoma (HNSCC) is crucial for effective treatment, particularly in metastatic cases. In this study, we analyzed multicenter bulk sequencing and comprehensive single-cell data from 702,446 cells, leading to the identification of a novel subtype of cancer-associated fibroblasts (CAFs), termed Secreted Frizzled-Related Protein2 CAFs (SFRP2_CAFs). These cells, originating from smooth muscle cells, display unique characteristics resembling both myofibroblastic CAFs and inflammatory CAFs, and are linked to poorer survival outcomes in HNSCC patients. Our findings reveal significant interactions between SFRP2_CAFs and SPP1 tumor-associated macrophages, which facilitate tumor invasion and metastasis. Moreover, our research identifies Nuclear factor I/X (NFIX) as a key transcription factor regulating SFRP2_CAFs behavior, confirmed through gene regulatory network analysis and simulation perturbation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

