HES1 potentiates high salt stress response as an enhancer of NFAT5-DNA binding
- PMID: 39384976
- PMCID: PMC11464898
- DOI: 10.1038/s42003-024-06997-7
HES1 potentiates high salt stress response as an enhancer of NFAT5-DNA binding
Abstract
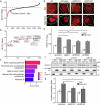
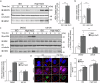
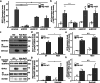
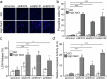
High salt conditions and subsequent hyperosmolarity are injurious cellular stresses that can activate immune signaling. Nuclear factor of activated T-cells 5 (NFAT5) is an essential transcription factor that induces osmoprotective genes such as aldose reductase (AR) and betaine-GABA transporter 1 (BGT1). High salt stress-mediated NFAT5 activation is also reported to accelerate the inflammatory response and autoimmune diseases. However, the systemic regulation of NFAT5 remains unclear. Here, we performed a genome-wide siRNA screen to comprehensively identify the regulators of NFAT5. We monitored NFAT5 nuclear translocation and identified one of the Notch signaling effectors, Hairy and enhancer of split-1 (HES1), as a positive regulator of NFAT5. HES1 was induced by high salinity via ERK signaling and facilitated NFAT5 recruitment to its target promoter region, resulting in the proper induction of osmoprotective genes and cytoprotection under high salt stress. These findings suggest that, though HES1 is well known as a transcriptional repressor, it positively regulates NFAT5-dependent transcription in the context of a high salinity/hyperosmotic response.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- JP21gm5010001/Japan Agency for Medical Research and Development (AMED)
- JP24gm0010009/Japan Agency for Medical Research and Development (AMED)
- JP17H06419/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP18H03995/MEXT | Japan Society for the Promotion of Science (JSPS)
- JP21H04760/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

