One-step nanoscale expansion microscopy reveals individual protein shapes
- PMID: 39385007
- PMCID: PMC7616833
- DOI: 10.1038/s41587-024-02431-9
One-step nanoscale expansion microscopy reveals individual protein shapes
Abstract
The attainable resolution of fluorescence microscopy has reached the subnanometer range, but this technique still fails to image the morphology of single proteins or small molecular complexes. Here, we expand the specimens at least tenfold, label them with conventional fluorophores and image them with conventional light microscopes, acquiring videos in which we analyze fluorescence fluctuations. One-step nanoscale expansion (ONE) microscopy enables the visualization of the shapes of individual membrane and soluble proteins, achieving around 1-nm resolution. We show that conformational changes are readily observable, such as those undergone by the ~17-kDa protein calmodulin upon Ca2+ binding. ONE is also applied to clinical samples, analyzing the morphology of protein aggregates in cerebrospinal fluid from persons with Parkinson disease, potentially aiding disease diagnosis. This technology bridges the gap between high-resolution structural biology techniques and light microscopy, providing new avenues for discoveries in biology and medicine.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: S.O.R. and F.O. are shareholders of NanoTag Biotechnologies GmbH. E.S.B. is an inventor on multiple patents related to ExM and co-founder of a company working on commercial applications of ExM. The other authors declare no competing interests. Ethics statement: Animals (Wistar rats, P0–P1) were treated according to the regulations of the local authority, the Lower Saxony State Office for Consumer Protection and Food Safety (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit), under the license Tötungsversuch T09/08. The informed consent of all of participants was obtained at the Paracelsus Elena Klinik, following the principles of the Declaration of Helsinki.
Figures

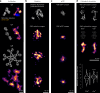

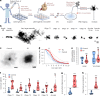
References
-
- Sahl, S. J. et al. Direct optical measurement of intra-molecular distances down to the Ångström scale. Preprint at bioRxiv10.1101/2023.07.07.548133 (2023).
MeSH terms
Substances
Grants and funding
- R01 AG070831/AG/NIA NIH HHS/United States
- MC_EX_MR/T046279/1/MRC_/Medical Research Council/United Kingdom
- MC_UP_1201/15/MRC_/Medical Research Council/United Kingdom
- MR/L009609/1/RCUK | Medical Research Council (MRC)
- SFB894/A9/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB1286/A03/B02/C06/Z04/Deutsche Forschungsgemeinschaft (German Research Foundation)
- MR/L009609/1/RCUK | MRC | Medical Research Foundation
- MC_EX_MR/T046279/1/RCUK | Medical Research Council (MRC)
- MR/L009609/1/MRC_/Medical Research Council/United Kingdom
- MC_UP_1201/15/RCUK | Medical Research Council (MRC)
- R01 EB024261/EB/NIBIB NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

