Detection and characterization of Campylobacter in air samples from poultry houses using shot-gun metagenomics - a pilot study
- PMID: 39385092
- PMCID: PMC11462905
- DOI: 10.1186/s12866-024-03563-3
Detection and characterization of Campylobacter in air samples from poultry houses using shot-gun metagenomics - a pilot study
Abstract
Background: Foodborne pathogens such as Campylobacter jejuni are responsible for a large proportion of the gastrointestinal infections worldwide associated with poultry meat. Campylobacter spp. can be found in the chicken fecal microbiome and can contaminate poultry meat during the slaughter process. Commonly used sampling methods to detect Campylobacter spp. at poultry farms use fecal droppings or boot swabs in combination with conventional culture techniques or PCR. In this pilot study, we have used air filtering and filters spiked with mock communities in combination with shotgun metagenomics to detect Campylobacter and test the applicability of this approach for the detection and characterization of foodborne pathogens. To the best of our knowledge is this the first study that combines air filtering with shotgun metagenomic sequencing for detection and characterization of Campylobacter.
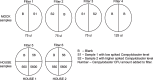
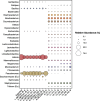
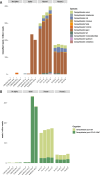
Results: Analysis of air filters spiked with different levels of Campylobacter, into a background of mock or poultry house communities, indicated that we could detect as little as 200 colony forming units (CFU) Campylobacter per sample using our protocols. The results indicate that even with limited sequencing effort we could detect Campylobacter in the samples analysed in this study. We observed significant amounts of Campylobacter in real-life samples from poultry houses using both real-time PCR as well as shotgun metagenomics, suggesting that the flocks in both houses were infected with Campylobacter spp. Interestingly, in both houses we find diverse microbial communities present in the indoor air which reflect the fecal microbiome of poultry. Some of the identified genera such as Staphylococcus, Escherichia and Pseudomonas are known to contain opportunistic pathogenic species.
Conclusions: These results show that air sampling of poultry houses in combination with shotgun metagenomics can detect and identify Campylobacter spp. present at low levels. This is important since early detection of Campylobacter enables measures to be put in place to ensure the safety of broiler products, animal health and public health. This approach has the potential to detect any pathogen present in poultry house air.
Keywords: Campylobacter spp.; Gelatin air filter; Metagenomics; Microbial communities; Mock communities; Spike controls.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- The global view of campylobacteriosis. Expert consulation. Geneva: World Health Organization; 2013.
-
- EFSA, ECDC. European Food Safety Authority & European Centre for Disease Prevention and Control. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food‐borne outbreaks in 2014. EFSA J. 2015;13. 10.2903/j.efsa.2015.4329.
-
- EFSA Panel on Biological Hazards (BIOHAZ). Scientific opinion on quantification of the risk posed by broiler meat to human campylobacteriosis in the EU. EFSA J. 2010;8: 1437. 10.2903/j.efsa.2010.1437. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

