DNA demethylation triggers cell free DNA release in colorectal cancer cells
- PMID: 39385243
- PMCID: PMC11462661
- DOI: 10.1186/s13073-024-01386-5
DNA demethylation triggers cell free DNA release in colorectal cancer cells
Abstract
Background: Liquid biopsy based on cell-free DNA (cfDNA) analysis holds significant promise as a minimally invasive approach for the diagnosis, genotyping, and monitoring of solid malignancies. Human tumors release cfDNA in the bloodstream through a combination of events, including cell death, active and passive release. However, the precise mechanisms leading to cfDNA shedding remain to be characterized. Addressing this question in patients is confounded by several factors, such as tumor burden extent, anatomical and vasculature barriers, and release of nucleic acids from normal cells. In this work, we exploited cancer models to dissect basic mechanisms of DNA release.
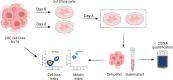
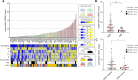
Methods: We measured cell loss ratio, doubling time, and cfDNA release in the supernatant of a colorectal cancer (CRC) cell line collection (N = 76) representative of the molecular subtypes previously identified in cancer patients. Association analyses between quantitative parameters of cfDNA release, cell proliferation, and molecular features were evaluated. Functional experiments were performed to test the impact of modulating DNA methylation on cfDNA release.
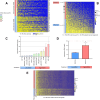
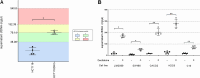
Results: Higher levels of supernatant cfDNA were significantly associated with slower cell cycling and increased cell death. In addition, a higher cfDNA shedding was found in non-CpG Island Methylator Phenotype (CIMP) models. These results indicate a positive correlation between lower methylation and increased cfDNA levels. To explore this further, we exploited methylation microarrays to identify a subset of probes significantly associated with cfDNA shedding and derive a methylation signature capable of discriminating high from low cfDNA releasers. We applied this signature to an independent set of 176 CRC cell lines and patient derived organoids to select 14 models predicted to be low or high releasers. The methylation profile successfully predicted the amount of cfDNA released in the supernatant. At the functional level, genetic ablation of DNA methyl-transferases increased chromatin accessibility and DNA fragmentation, leading to increased cfDNA release in isogenic CRC cell lines. Furthermore, in vitro treatment of five low releaser CRC cells with a demethylating agent was able to induce a significant increase in cfDNA shedding.
Conclusions: Methylation status of cancer cell lines contributes to the variability of cfDNA shedding in vitro. Changes in methylation pattern are associated with cfDNA release levels and might be exploited to increase sensitivity of liquid biopsy assays.
Keywords: Cell cycle; Cell death; Colorectal cancer; DNA methylation; Liquid biopsy; MSI; cfDNA.
© 2024. The Author(s).
Conflict of interest statement
F.D.N. has received speaker’s fees from Illumina and Pierre Fabre outside of the submitted work. A.Bard. reports receiving grants/research support from Neophore, AstraZeneca, Boehringer Ingelheim, and honoraria/consultation fees from Guardant Health. A.B. is a stock shareholder of Neophore and Kither Biotech. A.B. is an advisory board member for Neophore. L.B. is currently an employee of Biocartis NV. D.O. is currently an employee of AstraZeneca. The remaining authors declare that they have no competing interests.
Figures
References
-
- Di Nicolantonio F, Vitiello PP, Marsoni S, Siena S, Tabernero J, Trusolino L, et al. Precision oncology in metastatic colorectal cancer — from biology to medicine. Vol. 18, Nature Reviews Clinical Oncology. Nature Research; 2021. p. 506–25. - PubMed
-
- P. Mandel PM. Nuclear Acids In Human Blood Plasma. C R Seances Soc Biol Fil. 1948; - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

