Mucin-driven ecological interactions in an in vitro synthetic community of human gut microbes
- PMID: 39385462
- PMCID: PMC11632381
- DOI: 10.1093/glycob/cwae085
Mucin-driven ecological interactions in an in vitro synthetic community of human gut microbes
Abstract
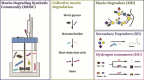
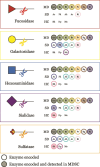
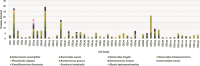
Specific human gut microbes inhabit the outer mucus layer of the gastrointestinal tract. Certain residents of this niche can degrade the large and complex mucin glycoproteins that constitute this layer and utilise the degradation products for their metabolism. In turn, this microbial mucin degradation drives specific microbiological ecological interactions in the human gut mucus layer. However, the exact nature of these interactions remains unknown. In this study, we designed and studied an in vitro mucin-degrading synthetic community that included mucin O-glycan degraders and cross-feeding microorganisms by monitoring community composition and dynamics through a combination of 16S rRNA gene amplicon sequencing and qPCR, mucin glycan degradation with PGC-LC-MS/MS, production of mucin-degrading enzymes and other proteins through metaproteomics, and metabolite production with HPLC. We demonstrated that specialist and generalist mucin O-glycan degraders stably co-exist and found evidence for cross-feeding relationships. Cross-feeding on the products of mucin degradation by other gut microbes resulted in butyrate production, hydrogenotrophic acetogenesis, sulfate reduction and methanogenesis. Metaproteomics analysis revealed that mucin glycan degraders Akkermansia muciniphila, Bacteroides spp. and Ruminococcus torques together contributed 92% of the total mucin O-glycan degrading enzyme pool of this community. Furthermore, comparative proteomics showed that in response to cultivation in a community compared to monoculture, mucin glycan degraders increased carbohydrate-active enzymes whereas we also found indications for niche differentiation. These results confirm the complexity of mucin-driven microbiological ecological interactions and the intricate role of carbohydrate-active enzymes in the human gut mucus layer.
Keywords: Akkermansia; CAZymes; gut microbiota; mucin; synthetic community.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures







References
-
- Acker L, Bergner KG, Diemair W, Heimann W, Kiermeier F, Schormüller J, Souci SW. Analytik der Lebensmittel, Nachweis und Bestimmung von Lebensmittel Inhaltsstoffen. In: Belitz H-D, Bergner K-G, Berndt D et al., editors. Handbuch der Lebensmittelchemie. Berlin, Germany: Springer Verlag; 1967.
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010.
-
- Apprill A, Mcnally S, Parsons R, Weber L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb Ecol. 2015:75(2):129–137.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

