Chromosomal integrons are genetically and functionally isolated units of genomes
- PMID: 39385642
- PMCID: PMC11551772
- DOI: 10.1093/nar/gkae866
Chromosomal integrons are genetically and functionally isolated units of genomes
Erratum in
-
Correction to 'Chromosomal integrons are genetically and functionally isolated units of genomes'.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf196. doi: 10.1093/nar/gkaf196. Nucleic Acids Res. 2025. PMID: 40052827 Free PMC article. No abstract available.
Abstract
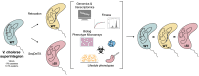
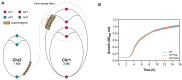
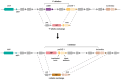
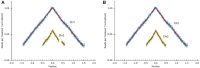
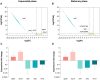
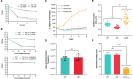
Integrons are genetic elements that increase the evolvability of bacteria by capturing new genes and stockpiling them in arrays. Sedentary chromosomal integrons (SCIs) can be massive and highly stabilized structures encoding hundreds of genes, whose function remains generally unknown. SCIs have co-evolved with the host for aeons and are highly intertwined with their physiology from a mechanistic point of view. But, paradoxically, other aspects, like their variable content and location within the genome, suggest a high genetic and functional independence. In this work, we have explored the connection of SCIs to their host genome using as a model the Superintegron (SI), a 179-cassette long SCI in the genome of Vibrio cholerae N16961. We have relocated and deleted the SI using SeqDelTA, a novel method that allows to counteract the strong stabilization conferred by toxin-antitoxin systems within the array. We have characterized in depth the impact in V. cholerae's physiology, measuring fitness, chromosome replication dynamics, persistence, transcriptomics, phenomics, natural competence, virulence and resistance against protist grazing. The deletion of the SI did not produce detectable effects in any condition, proving that-despite millions of years of co-evolution-SCIs are genetically and functionally isolated units of genomes.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Mazel D. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 2006; 4:608–620. - PubMed
-
- Escudero J., Loot C., Nivina A., Mazel D. The Integron: adaptation on demand. Microbiol. Spectr. 2015; 13:MDNA3–A0019–2014. - PubMed
-
- Stokes H.W., Hall R.M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol. Microbiol. 1989; 3:1669–1683. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

