Cyclin C promotes development and progression of B-cell acute lymphoblastic leukemia by counteracting p53-mediated stress responses
- PMID: 39385738
- PMCID: PMC11959249
- DOI: 10.3324/haematol.2024.285701
Cyclin C promotes development and progression of B-cell acute lymphoblastic leukemia by counteracting p53-mediated stress responses
Abstract
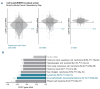
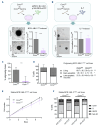
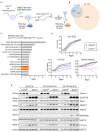
Despite major therapeutic advances in the treatment of acute lymphoblastic leukemia (ALL), resistances and long-term toxicities still pose significant challenges. Cyclins and their associated cyclin-dependent kinases are one focus of cancer research when looking for targeted therapies. We discovered cyclin C to be a key factor for B-cell ALL (B-ALL) development and maintenance. While cyclin C is not essential for normal hematopoiesis, CcncΔ/Δ BCR::ABL1+ B-ALL cells fail to elicit leukemia in mice. RNA sequencing experiments revealed a p53 pathway deregulation in CcncΔ/Δ BCR::ABL1+ cells resulting in the inability of the leukemic cells to adequately respond to stress. A genome-wide CRISPR/Cas9 loss-of-function screen supplemented with additional knock-outs unveiled a dependency of human B-lymphoid cell lines on CCNC. High cyclin C levels in B-cell precursor (BCP) ALL patients were associated with poor event-free survival and increased risk of early disease recurrence after remission. Our findings highlight cyclin C as a potential therapeutic target for B-ALL, particularly to enhance cancer cell sensitivity to stress and chemotherapy.
Figures




References
-
- Groffen J, Stephenson JR, Heisterkamp N, de Klein A, Bartram CR, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36(1):93-99. - PubMed
-
- Pui CH, Jeha S. New therapeutic strategies for the treatment of acute lymphoblastic leukaemia. Nat Rev Drug Discov. 2007;6(2):149-165. - PubMed
-
- Quintás-Cardama A, Kantarjian H, Cortes J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat Rev Drug Discov. 2007;6(10):834-848. - PubMed
-
- Lee H, Basso IN, Kim DDH. Target spectrum of the BCR-ABL tyrosine kinase inhibitors in chronic myeloid leukemia. Int J Hematol. 2021;113(5):632-641. - PubMed
-
- Yang K, Fu L-W. Mechanisms of resistance to BCR-ABL TKIs and the therapeutic strategies: a review. Crit Rev Oncol Hematol. 2015;93(3):277-292. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

