Phylogenetic evidence of extensive spatial mixing of diverse HIV-1 group M lineages within Cameroon but not between its neighbours
- PMID: 39386075
- PMCID: PMC11463025
- DOI: 10.1093/ve/veae070
Phylogenetic evidence of extensive spatial mixing of diverse HIV-1 group M lineages within Cameroon but not between its neighbours
Abstract
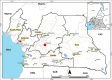
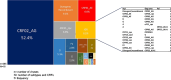
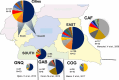
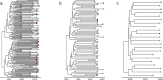
From the perspective of developing relevant interventions for treating HIV and controlling its spread, it is particularly important to comprehensively understand the underlying diversity of the virus, especially in countries where the virus has been present and evolving since the cross-species transmission event that triggered the global pandemic. Here, we generate and phylogenetically analyse sequences derived from the gag-protease (2010 bp; n = 115), partial integrase (345 bp; n = 36), and nef (719 bp; n = 321) genes of HIV-1 group M (HIV-1M) isolates sampled between 2000 and 2022 from two cosmopolitan cities and 40 remote villages of Cameroon. While 52.4% of all sequenced viruses belonged to circulating recombinant form (CRF) 02_AG (CRF02_AG), the remainder were highly diverse, collectively representing seven subtypes and sub-subtypes, eight CRFs, and 36 highly divergent lineages that fall outside the established HIV-1M classification. Additionally, in 77 samples for which at least two genes were typed, 31% of the studied viruses apparently had fragments from viruses belonging to different clades. Furthermore, we found that the distribution of HIV-1M populations is similar between different regions of Cameroon. In contrast, HIV-1M demographics in Cameroon differ significantly from those in its neighbouring countries in the Congo Basin (CB). In phylogenetic trees, viral sequences cluster according to the countries where they were sampled, suggesting that while there are minimal geographical or social barriers to viral dissemination throughout Cameroon, there is strongly impeded dispersal of HIV-1M lineages between Cameroon and other locations of the CB. This suggests that the apparent stability of highly diverse Cameroonian HIV-1M populations may be attributable to the extensive mixing of human populations within the country and the concomitant trans-national movements of major lineages with very similar degrees of fitness; coupled with the relatively infrequent inter-national transmission of these lineages from neighbouring countries in the CB.
Keywords: Cameroon; HIV-1 group M (HIV-1M); evolution; phylogenetic; viral diversity.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Centre de Documentation Numérique du Secteur Santé . Evaluation de l’impact du VIH sur la Population au Cameroun CAMPHIA 2017-2018. 2020. http://cdnss.minsante.cm/?q=fr/content/evaluation-de-l%E2%80%99impact-du... (accessed 7 July 2023).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

