Motif discovery in hospital ward vital signs observation networks
- PMID: 39386086
- PMCID: PMC11458707
- DOI: 10.1007/s13721-024-00490-1
Motif discovery in hospital ward vital signs observation networks
Abstract
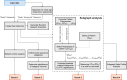
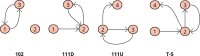
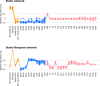
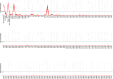
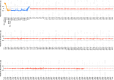
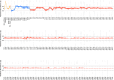
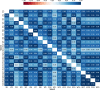
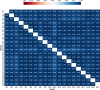
Vital signs observations are regular measurements used by healthcare staff to track a patient's overall health status on hospital wards. We look at the potential in re-purposing aggregated and anonymised hospital data sources surrounding vital signs recording to provide new insights into how care is managed and delivered on wards. In this paper, we conduct a retrospective longitudinal observational study of 770,720 individual vital signs recordings across 20 hospital wards in South Wales (UK) and present a network modelling framework to explore and extract behavioural patterns via analysis of the resulting network structures at a global and local level. Self-loop edges, dyad, triad, and tetrad subgraphs were extracted and evaluated against a null model to determine individual statistical significance, and then combined into ward-level feature vectors to provide the means for determining notable behaviours across wards. Modelling data as a static network, by aggregating all vital sign observation data points, resulted in high uniformity but with the loss of important information which was better captured when modelling the static-temporal network, highlighting time's crucial role as a network element. Wards mostly followed expected patterns, with chains or stand-alone supplementary observations by clinical staff. However, observation sequences that deviate from this are revealed in five identified motif subgraphs and 6 anti-motif subgraphs. External ward characteristics also showed minimal impact on the relative abundance of subgraphs, indicating a 'superfamily' phenomena that has been similarly seen in complex networks in other domains. Overall, the results show that network modelling effectively captured and exposed behaviours within vital signs observation data, and demonstrated uniformity across hospital wards in managing this practice.
Keywords: Motif discovery; Network analysis; Retrospective study; Subgraph ratio profile; Vital signs observations.
© The Author(s) 2024.
Conflict of interest statement
Conflict of interestAll authors declare they have no Conflict of interest.
Figures












References
-
- ABUHB (2017) Aneurin bevan university health board deteriorating patient policy
-
- Agasse-Duval M, Lawford S (2018) Subgraphs and motifs in a dynamic airline network. CoRR. arXiv:1807.02585
-
- Alon U (2003) Biological networks: the tinkerer as an engineer. Science 301:1866–1867. 10.1126/science.1089072 - PubMed
-
- Armstrong B, Walthall H, Clancy M et al (2008) Recording of vital signs in a district general hospital emergency department. Emerg Med J 25(12):799–802. 10.1136/emj.2007.052951 - PubMed
-
- Artzy-Randrup Y, Fleishman S, Ben-Tal N et al (2004) Comment on “network motifs: Simple building blocks of complex networks’’ and “superfamilies of evolved and designed networks’’. Science. 10.1126/science.1099334 - PubMed
LinkOut - more resources
Full Text Sources
