Predicting host species susceptibility to influenza viruses and coronaviruses using genome data and machine learning: a scoping review
- PMID: 39386249
- PMCID: PMC11462629
- DOI: 10.3389/fvets.2024.1358028
Predicting host species susceptibility to influenza viruses and coronaviruses using genome data and machine learning: a scoping review
Abstract
Introduction: Predicting which species are susceptible to viruses (i.e., host range) is important for understanding and developing effective strategies to control viral outbreaks in both humans and animals. The use of machine learning and bioinformatic approaches to predict viral hosts has been expanded with advancements in in-silico techniques. We conducted a scoping review to identify the breadth of machine learning methods applied to influenza and coronavirus genome data for the identification of susceptible host species.
Methods: The protocol for this scoping review is available at https://hdl.handle.net/10214/26112. Five online databases were searched, and 1,217 citations, published between January 2000 and May 2022, were obtained, and screened in duplicate for English language and in-silico research, covering the use of machine learning to identify susceptible species to viruses.
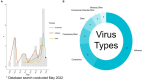
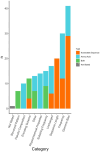
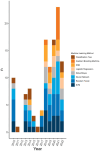
Results: Fifty-three relevant publications were identified for data charting. The breadth of research was extensive including 32 different machine learning algorithms used in combination with 29 different feature selection methods and 43 different genome data input formats. There were 20 different methods used by authors to assess accuracy. Authors mostly used influenza viruses (n = 31/53 publications, 58.5%), however, more recent publications focused on coronaviruses and other viruses in combination with influenza viruses (n = 22/53, 41.5%). The susceptible animal groups authors most used were humans (n = 57/77 analyses, 74.0%), avian (n = 35/77 45.4%), and swine (n = 28/77, 36.4%). In total, 53 different hosts were used and, in most publications, data from multiple hosts was used.
Discussion: The main gaps in research were a lack of standardized reporting of methodology and the use of broad host categories for classification. Overall, approaches to viral host identification using machine learning were diverse and extensive.
Keywords: coronaviruses; genome; influenza A viruses; interspecies transmission; machine-learning; scoping review; spillover.
Copyright © 2024 Alberts, Berke, Rocha, Keay, Maboni and Poljak.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Miller RS, Sweeney SJ, Slootmaker C, Grear DA, Di Salvo PA, Kiser D, et al. . Cross-species transmission potential between wild pigs, livestock, poultry, wildlife, and humans: implications for disease risk Management in North America. Sci Rep. (2017) 7:7821. doi: 10.1038/s41598-017-07336-z - DOI - PMC - PubMed
-
- Claes F, Kuznetsov D, Liechti R, Von Dobschuetz S, Dinh Truong B, Gleizes A, et al. . The EMPRES-i genetic module: a novel tool linking epidemiological outbreak information and genetic characteristics of influenza viruses. Database. (2014) 2014:bau008. doi: 10.1093/database/bau008 - DOI - PMC - PubMed
-
- Fermin G. Chapter 5 - Host Range, Host–Virus Interactions, and Virus Transmission. In: P Tennant, G Fermin, JE Foster, editors. Viruses [Internet]. London, United Kingdom: Academic Press; (2018). p. 101–34. Available from: https://www.sciencedirect.com/science/article/pii/B978012811257100005X.
Publication types
LinkOut - more resources
Full Text Sources

