Unveiling the Molecular Features of SCLC With a Clinical RNA Expression Panel
- PMID: 39386315
- PMCID: PMC11459576
- DOI: 10.1016/j.jtocrr.2024.100723
Unveiling the Molecular Features of SCLC With a Clinical RNA Expression Panel
Abstract
Introduction: The translation of gene expression profiles of SCLC to clinical testing remains relatively unexplored. In this study, gene expression variations in SCLC were evaluated to identify potential biomarkers.
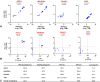
Methods: RNA expression profiling was performed on 44 tumor samples from 35 patients diagnosed with SCLC using the clinically validated RNA Salah Targeted Expression Panel (RNA STEP). RNA sequencing (RNA-Seq) and immunohistochemistry were performed on two different SCLC cohorts, and correlation analyses were performed for the ASCL1, NEUROD1, POU2F3, and YAP1 genes and their corresponding proteins. RNA STEP and RNA-Seq results were evaluated for gene expression profiles and heterogeneity between SCLC primary and metastatic sites. RNA STEP gene expression profiles of independent SCLC samples (n = 35) were compared with lung adenocarcinoma (n = 160) and squamous cell carcinoma results (n = 25).
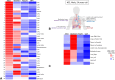
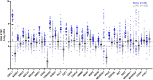
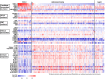
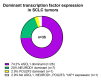
Results: The RNA STEP results were highly correlated with RNA-Seq and immunohistochemistry results. The dominant transcription regulator by RNA STEP was ASCL1 in 74.2% of the samples, NEUROD1 in 20%, and POU2F3 in 2.9%. The ASCL1, NEUROD1, and POU2F3 gene expression profiles were heterogeneous between primary and metastatic sites. SCLCs displayed markedly high expression for targetable genes DLL3, EZH2, TERT, and RET. SCLCs were found to have relatively colder immune profiles than lung adenocarcinomas and squamous cell carcinomas, characterized by lower expression of HLA genes, immune cell, and immune checkpoint genes, except the LAG3 gene.
Conclusions: Clinical-grade SCLC RNA expression profiling has value for SCLC subtyping, design of clinical trials, and identification of patients for trials and potential targeted therapy.
Keywords: Biomarker; Clinical testing; RNA expression; Small cell lung cancer; Transcriptomics.
© 2024 The Authors.
Conflict of interest statement
Dr. Chiappori received funding from 10.13039/100002491Bristol-Myers Squibb for the clinical trial (MCC19163). Dr. Boyle and Dr. Koomen declare grants/contracts with Bristol-Myers Squibb unrelated to this research. Dr. Perez declares receiving grants from Bristol-Myers Squibb; providing consulting services for AstraZeneca, Bristol-Myers Squibb, G1 Therapeutics, and Novocure; and having board membership in Out of Zion. Dr. Haura declares providing consulting services for Kanaph Therapeutics and ORI Capital II; receiving research funding from 10.13039/100019364Revolution Medicines; and providing advisory services for RevMed and Janssen, all unrelated to this research. The remaining authors declare no conflict of interest.
Figures





References
-
- College of American Pathologists. The CAP cancer protocols. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/canc.... Accessed July 2024.
-
- Cedrés S., Ponce-Aix S., Pardo-Aranda N., et al. Analysis of expression of PTEN/PI3K pathway and programmed cell death ligand 1 (PD-L1) in malignant pleural mesothelioma (MPM) Lung Cancer. 2016;96:1–6. - PubMed
-
- Fujino K., Motooka Y., Hassan W.A., et al. Insulinoma-associated Protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol. 2015;185:3164–3177. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

