This is a preprint.
Discovery of electrophilic degraders that exploit SNAr chemistry
- PMID: 39386645
- PMCID: PMC11463635
- DOI: 10.1101/2024.09.25.615094
Discovery of electrophilic degraders that exploit SNAr chemistry
Abstract
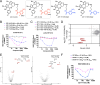
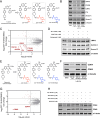
Targeted covalent inhibition (TCI) and targeted protein degradation (TPD) have proven effective in pharmacologically addressing formerly 'undruggable' targets. Integration of both methodologies has resulted in the development of electrophilic degraders where recruitment of a suitable E3 ubiquitin ligase is achieved through formation of a covalent bond with a cysteine nucleophile. Expanding the scope of electrophilic degraders requires the development of electrophiles with tempered reactivity that enable selective ligase recruitment and reduce cross-reactivity with other cellular nucleophiles. In this study, we report the use of chemical moieties that enable nucleophilic aromatic substitution (SNAr) reactions in the rational design of electrophilic protein degraders. Appending an SNAr covalent warhead to several preexisting small molecule inhibitors transformed them into degraders, obviating the need for a defined E3 ligase recruiter. The SNAr covalent warhead is versatile; it can recruit various E3 ligases, including DDB1 and CUL4 associated factor 11 (DCAF11), DDB1 and CUL4 associated factor 16 (DCAF16), and possibly others. The incorporation of an SNAr covalent warhead into the BRD4 inhibitor led to the discovery of degraders with low picomolar degradation potency. Furthermore, we demonstrate the broad applicability of this approach through rational functional switching from kinase inhibitors into potent degraders.
Conflict of interest statement
N.S.G. is a founder, science advisory board member (SAB) and equity holder in Syros, C4, Allorion, Lighthorse, Voronoi, Inception, Matchpoint, CobroVentures, GSK, Shenandoah (board member), Larkspur (board member) and Soltego (board member). The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Jansen, Kinogen, Arbella, Deerfield, Springworks, Interline and Sanofi. B.L.E. has received research funding from Novartis and Calico. He has received consulting fees from Abbvie. He is a member of the scientific advisory board and shareholder for Neomorph Inc., TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics. E.S.F. is a founder, scientific advisory board (SAB) member, and equity holder of Civetta Therapeutics, Proximity Therapeutics, and Neomorph, Inc. (also board of directors). He is an equity holder and SAB member for Avilar Therapeutics, Photys Therapeutics, and Ajax Therapeutics and an equity holder in Lighthorse Therapeutics. E.S.F. is a consultant to Novartis, EcoR1 capital, Odyssey and Deerfield. The Fischer lab receives or has received research funding from Deerfield, Novartis, Ajax, Interline, Bayer and Astellas. K.A.D. receives or has received consulting fees from Kronos Bio and Neomorph Inc. M.S. has received research funding from Calico Life Sciences LLC. All other authors declare no competing interests.
Figures


References
-
- Guedeney N., Cornu M., Schwalen F., Kieffer C. & Voisin-Chiret A.S. PROTAC technology: A new drug design for chemical biology with many challenges in drug discovery. Drug Discov Today 28, 103395 (2023). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
