The DNA Damage Repair Function of Fission Yeast CK1 Involves Targeting Arp8, a Subunit of the INO80 Chromatin Remodeling Complex
- PMID: 39387272
- PMCID: PMC11583621
- DOI: 10.1080/10985549.2024.2408016
The DNA Damage Repair Function of Fission Yeast CK1 Involves Targeting Arp8, a Subunit of the INO80 Chromatin Remodeling Complex
Abstract
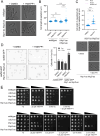
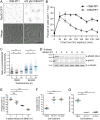
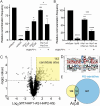
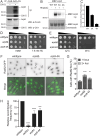
The CK1 family are conserved serine/threonine kinases with numerous substrates and cellular functions. The fission yeast CK1 orthologues Hhp1 and Hhp2 were first characterized as regulators of DNA repair, but the mechanism(s) by which CK1 activity promotes DNA repair had not been investigated. Here, we found that deleting Hhp1 and Hhp2 or inhibiting CK1 catalytic activities in yeast or in human cells increased double-strand breaks (DSBs). The primary pathways to repair DSBs, homologous recombination and nonhomologous end joining, were both less efficient in cells lacking Hhp1 and Hhp2 activity. To understand how Hhp1 and Hhp2 promote DNA damage repair, we identified new substrates of these enzymes using quantitative phosphoproteomics. We confirmed that Arp8, a component of the INO80 chromatin remodeling complex, is a bona fide substrate of Hhp1 and Hhp2 important for DNA repair. Our data suggest that Hhp1 and Hhp2 facilitate DNA repair by phosphorylating multiple substrates, including Arp8.
Keywords: Arp8; CK1, casein kinase 1; DNA repair; INO80; Schizosaccharomyces pombe; homologous recombination; nonhomologous end joining; phosphoproteomics.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
