Loss of Carbamoyl Phosphate Synthetase 1 Potentiates Hepatocellular Carcinoma Metastasis by Reducing Aspartate Level
- PMID: 39387452
- PMCID: PMC11615744
- DOI: 10.1002/advs.202402703
Loss of Carbamoyl Phosphate Synthetase 1 Potentiates Hepatocellular Carcinoma Metastasis by Reducing Aspartate Level
Abstract
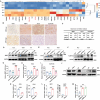
Hepatocellular carcinoma (HCC) is one of the most lethal cancers worldwide. Numerous studies have shown that metabolic reprogramming is crucial for the development of HCC. Carbamoyl phosphate synthase 1 (CPS1), a rate-limiting enzyme in urea cycle, is an abundant protein in normal hepatocytes, however, lacking systemic research in HCC. It is found that CPS1 is low-expressed in HCC tissues and circulating tumor cells, negatively correlated with HCC stage and prognosis. Further study reveals that CPS1 is a double-edged sword. On the one hand, it inhibits the activity of phosphatidylcholine-specific phospholipase C to block the biosynthesis of diacylglycerol (DAG), leading to the downregulation of the DAG/protein kinase C pathway to inhibit invasion and metastasis of cancer cells. On the other hand, CPS1 promotes cell proliferation by increasing intracellular S-adenosylmethionin to enhance the m6A modification of solute carrier family 1 member 3 mRNA, a key transporter for aspartate intake. Finally, CPS1 overexpressing adeno-associated virus can dampen HCC progression. Collectively, this results uncovered that CPS1 is a switch between HCC proliferation and metastasis by increasing intracellular aspartate level.
Keywords: CPS1; PC‐PLC; aspartate; m6A; metastasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Villanueva A., New Engl. J. Med. 2019, 380, 1450.
-
- Lu Y., Chan Y.‐T., Wu J., Feng Z., Yuan H., Li Q., Xing T., Xu L., Zhang C., Tan H.‐Y., Lee T. K.‐W., Feng Y., Wang N., Drug Resist. Update 2023, 71, 101015. - PubMed
-
- Wang S., Zhou L., Ji N., Sun C., Sun L., Sun J., Du Y., Zhang N., Li Y., Liu W., Lu W., Drug Resist. Update 2023, 69, 100976. - PubMed
-
- Li Q., Zhang L., Yang Q., Li M., Pan X., Xu J., Zhong C., Yao F., Zhang R., Zhou S., Dai X., Shi X., Dai Y., Xu J., Cheng X., Xiao W., She Z., Wang K., Qian X., Pu L., Zhang P., Wang X., Cell Metab. 2023, 35, 912. - PubMed
-
- Sun J., Ding J., Shen Q., Wang X., Wang M., Huang Y., Zhang X., Zhu H., Zhang F., Wu D., Peng M., Zhang Z., Yuan Y., Li W., She Z.‐G., Zhang X.‐J., Li H., Zhang P., Huang Z., J. Hepatol. 2023, 78, 627. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 82203791/National Natural Science Foundation of China
- 8237300/National Natural Science Foundation of China
- 2020jsz604/Open Project of Key Laboratory of Tumor Immunopathology, Ministry of Education
- Young and Middle-aged Senior Medical Talents Studio of Chongqing
- 13-004-009/Senior Medical Talents Program of Chongqing for Young, and Middle-aged
- CSTB2022NSCQ-MSX1010/Natural Science Foundation of Chongqing Municipality
- CSTB2022NSCQ-MSX1038/Natural Science Foundation of Chongqing Municipality
- CSTB2022NSCQ-MSX0775/Natural Science Foundation of Chongqing Municipality
- CSTB2022NSCQ-MSX1010/2.Chongqing Natural Science Foundation
- CSTB2022NSCQ-MSX1038/2.Chongqing Natural Science Foundation
- CSTB2022NSCQ-MSX0775/2.Chongqing Natural Science Foundation
- 13-004-009/Kuanren Talents Program of the Second Affiliated Hospital of Chongqing Medical University
LinkOut - more resources
Full Text Sources
Medical
