Rbm3 deficiency leads to transcriptome-wide splicing alterations
- PMID: 39387568
- PMCID: PMC11575738
- DOI: 10.1080/15476286.2024.2413820
Rbm3 deficiency leads to transcriptome-wide splicing alterations
Abstract
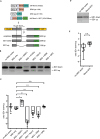
Rbm3 (RNA-binding motif protein 3) is a stress responsive gene, which maintains cellular homeostasis and promotes survival upon various harmful cellular stimuli. Rbm3 protein shows conserved structural and molecular similarities to heterogeneous nuclear ribonucleoproteins (hnRNPs), which regulate all steps of the mRNA metabolism. Growing evidence is pointing towards a broader role of Rbm3 in various steps of gene expression. Here, we demonstrate that Rbm3 deficiency is linked to transcriptome-wide pre-mRNA splicing alterations, which can be reversed through Rbm3 co-expression from a cDNA. Using an MS2 tethering assay, we show that Rbm3 regulates splice site selection similar to other hnRNP proteins when recruited between two competing 5 splice sites. Furthermore, we show that the N-terminal part of Rbm3 encompassing the RNA recognition motif (RRM), is sufficient to elicit changes in splice site selection. On the basis of these findings, we propose a novel, undescribed function of Rbm3 in RNA splicing that contributes to the preservation of transcriptome integrity.
Keywords: RNA-seq; Rbm3; isoform switch; splicing; transcriptional fidelity.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
