High-throughput screening for small-molecule stabilizers of misfolded glucocerebrosidase in Gaucher disease and Parkinson's disease
- PMID: 39388267
- PMCID: PMC11494340
- DOI: 10.1073/pnas.2406009121
High-throughput screening for small-molecule stabilizers of misfolded glucocerebrosidase in Gaucher disease and Parkinson's disease
Abstract
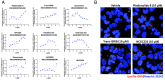
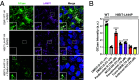
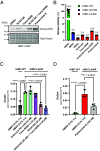
Glucocerebrosidase (GCase) is implicated in both a rare, monogenic disorder (Gaucher disease, GD) and a common, multifactorial condition (Parkinson's disease, PD); hence, it is an urgent therapeutic target. To identify correctors of severe protein misfolding and trafficking obstruction manifested by the pathogenic L444P-variant of GCase, we developed a suite of quantitative, high-throughput, cell-based assays. First, we labeled GCase with a small proluminescent HiBiT peptide reporter tag, enabling quantitation of protein stabilization in cells while faithfully maintaining target biology. TALEN-based gene editing allowed for stable integration of a single HiBiT-GBA1 transgene into an intragenic safe-harbor locus in GBA1-knockout H4 (neuroglioma) cells. This GD cell model was amenable to lead discovery via titration-based quantitative high-throughput screening and lead optimization via structure-activity relationships. A primary screen of 10,779 compounds from the NCATS bioactive collections identified 140 stabilizers of HiBiT-GCase-L444P, including both pharmacological chaperones (ambroxol and noninhibitory chaperone NCGC326) and proteostasis regulators (panobinostat, trans-ISRIB, and pladienolide B). Two complementary high-content imaging-based assays were deployed to triage hits: The fluorescence-quenched substrate LysoFix-GBA captured functional lysosomal GCase activity, while an immunofluorescence assay featuring antibody hGCase-1/23 directly visualized GCase lysosomal translocation. NCGC326 was active in both secondary assays and completely reversed pathological glucosylsphingosine accumulation. Finally, we tested the concept of combination therapy by demonstrating synergistic actions of NCGC326 with proteostasis regulators in enhancing GCase-L444P levels. Looking forward, these physiologically relevant assays can facilitate the identification, pharmacological validation, and medicinal chemistry optimization of small molecules targeting GCase, ultimately leading to a viable therapeutic for GD and PD.
Keywords: Gaucher disease; Parkinson’s disease; glucocerebrosidase; high-throughput screening; pharmacological chaperone.
Conflict of interest statement
Competing interests statement:A.G. and R.J. are employees of F. Hoffmann-La Roche AG.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

