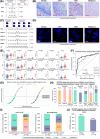
FIGURE 1
The development and validation of the QCIGISH model for cervical cancer risk stratification. (A) Schematic diagram of the QCIGISH technology as applied on bright field microscopy images. Blue components in the image are cell nuclei while red dots represent the activated gene loci. More red dots indicate aberrant allelic expression. N0, N1, N2, and N3+ refer to the total count of cell nuclei observed with 0, 1, 2, and 3 or more dots, respectively, with N0 and N1 collectively representing normal allelic expression, while N2 and N3+ both indicating aberrant allelic expression. BAE, MAE, and TE were computed by applying the values determined for N0, N1, N2 and N3+ using the given equations. Higher values for BAE, MAE, and TE indicate elevated epigenetic imprinting alterations. (B) Representative images of bright field QCIGISH detection results for benign, CIN1, CIN3, and malignant cervical tissue specimens showing a generally increasing allelic expression quantified using BAE, MAE, and TE measurements. (C) Schematic diagram of the QCIGISH technology as applied on fluorescent microscopy images. Blue components in the image are cell nuclei while green dots represent the activated gene loci. More green dots indicate aberrant allelic expression. N0, N1, N2, N3, N4, N5, N6, N7, N8, and N9+ refer to the total count of cell nuclei observed with 0, 1, 2, 3, 4, 5, 6, 7, 8, and 9 or more dots, respectively, with N0 and N1 collectively representing normal allelic expression, while N2, N3, N4, N5, N6, N7, N8, and N9+ all indicating aberrant allelic expression. BAE, MAE3‐4, MAE5‐6, MAE7‐8, MAE9+, and TE were computed with the values determined for N0, N1, N2, N3, N4, N5, N6, N7, N8 and N9+ using the given equations. Higher values for BAE, MAE3‐4, MAE5‐6, MAE7‐8, MAE9+, and TE indicate higher epigenetic imprinting alterations. (D) Representative images of fluorescent QCIGISH detection results for benign, CIN1, CIN3, and malignant cervical cytological specimens showing a generally increasing allelic expression quantified using BAE, MAE3‐4, MAE5‐6, MAE7‐8, MAE9+, and TE measurements. (E) Elevated epigenetic imprinting alterations in terms of BAE, MAE3‐4, MAE5‐6, MAE7‐8, MAE9+, and TE demonstrated for the combined CIN3 and malignant categories as compared to cases diagnosed as benign and CIN1 for the GNAS, HM13 and SNU13 imprinted genes. Robust Rank‐Order Test was applied during statistical evaluation. *P < 0.05. **P < 0.01. ***P < 0.001. ns, not significant. (F) Improved malignancy discrimination performance in terms of the AUC demonstrated for the individual gene models employing the top four best performing biomarker features, and the final combined gene model utilizing weights equal to 40%, 40%, and 20% for the GNAS, HM13 and SNU13 imprinted genes, respectively, as compared to the individual gene models employing the top four best performing biomarker features. (G) Logistic curve plot of the estimated cervical cancer probabilities for the final combined gene model as evaluated on the model validation set showing sufficient discrimination between benign and CIN1 cases against CIN3 and malignant cases. (H) Diagnostic performance in the model validation set showing 93.8% sensitivity (95% CI 85.4%‐100.0%) on combined CIN3 and malignant cases, 83.60% specificity (95% CI 75.1%‐92.1%) on combined benign and CIN1 cases, 71.4% PPV (95% CI 57.8%‐85.1%) on combined CIN3 and malignant cases, and 96.8% NPV (95% CI 92.5%‐100.0%) on combined benign and CIN1 cases. Wald's 95% confidence intervals determined using normal approximation were used. (I) Triage performance of the QCIGISH diagnostic model on two groups of hrHPV‐positive patients: (1) detected with 16/18 genotypes and (2) detected with non‐16/18 genotypes. (J) Triage performance of the QCIGISH diagnostic model on two combined groups of patients who are clinically recommended for colposcopy: (1) hrHPV‐positive for 16/18 genotypes; and (2) hrHPV‐positive for other genotypes but with TCT grades evaluated as atypical squamous cells of undetermined significance and above (ASCUS+). Group sample sizes are represented as n. †Cases with hrHPV16/18, regardless of the TCT diagnosis, were included in the analysis. The ten cases detected as hrHPV‐positive but with indeterminate genotype were all diagnosed as TCT ASCUS+ and were included in the analysis. Abbreviations: QCIGISH, Quantitative chromogenic imprinted gene in‐situ hybridization; CIN, Cervical intraepithelial neoplasia; BAE, Bi‐allelic expression; MAE, Multi‐allelic expression; TE, Total expression; GNAS, Guanine nucleotide‐binding protein, alpha‐stimulating complex locus; HM13, Histocompatibility minor 13; SNU13, Small nuclear ribonucleoprotein 13; AUC, Area under the receiver operating characteristic curve; CI, confidence interval; HPV, Human papillomavirus; TCT, Thinprep cytologic test; ASCUS, Atypical squamous cells of undetermined significance; hrHPV, High‐risk human papillomavirus.