A gene desert required for regulatory control of pleiotropic Shox2 expression and embryonic survival
- PMID: 39389973
- PMCID: PMC11467299
- DOI: 10.1038/s41467-024-53009-7
A gene desert required for regulatory control of pleiotropic Shox2 expression and embryonic survival
Abstract
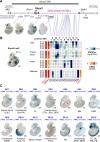
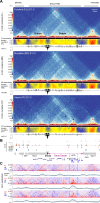
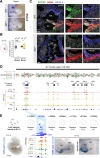
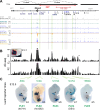
Approximately a quarter of the human genome consists of gene deserts, large regions devoid of genes often located adjacent to developmental genes and thought to contribute to their regulation. However, defining the regulatory functions embedded within these deserts is challenging due to their large size. Here, we explore the cis-regulatory architecture of a gene desert flanking the Shox2 gene, which encodes a transcription factor indispensable for proximal limb, craniofacial, and cardiac pacemaker development. We identify the gene desert as a regulatory hub containing more than 15 distinct enhancers recapitulating anatomical subdomains of Shox2 expression. Ablation of the gene desert leads to embryonic lethality due to Shox2 depletion in the cardiac sinus venosus, caused in part by the loss of a specific distal enhancer. The gene desert is also required for stylopod morphogenesis, mediated via distributed proximal limb enhancers. In summary, our study establishes a multi-layered role of the Shox2 gene desert in orchestrating pleiotropic developmental expression through modular arrangement and coordinated dynamics of tissue-specific enhancers.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Craig Venter, J. et al. The Sequence of the Human Genome. Science291, 1304–1351 (2001). - PubMed
-
- Nobrega, M. A., Ovcharenko, I., Afzal, V. & Rubin, E. M. Scanning human gene deserts for long-range enhancers. Science302, 413 (2003). - PubMed
-
- Nóbrega, M. A., Zhu, Y., Plajzer-Frick, I., Afzal, V. & Rubin, E. M. Megabase deletions of gene deserts result in viable mice. Nature431, 984–988 (2004). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R01 HG003988/HG/NHGRI NIH HHS/United States
- RGPIN-2019-04812/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- UM1 HL098166/HL/NHLBI NIH HHS/United States
- U54 HG006997/HG/NHGRI NIH HHS/United States
- R24 HL123879/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

